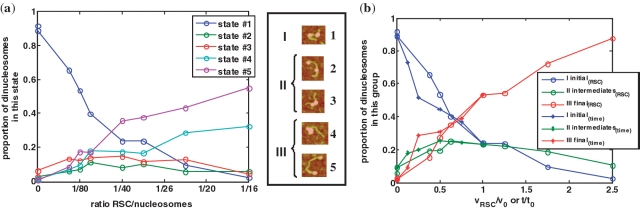
Figure 3.
Experimental dinucleosome populations in each state: titration and time evolution. (a) Evolution of the proportion of dinucleosomes in each State (#1–#5) during the sliding reaction by RSC. Nucleosome populations are counted from analysis of AFM images obtained for various RSC/nucleosome ratio and a fixed incubation time of 60 min at 29°C (the RSC/nucleosome ratio is estimated of approximately 1/40 for v0 = 0.4 µl). (b) The proportion of dinucleosomes in each group of states (I initial, II intermediate and III final) is reported as a function of general reaction coordinates: RSC volume (circles) or time (stars). The x-axis has been normalized by v0 = 0.4 µl of RSC for the titration and τ = 60 min for the kinetics. For each experimental data points, about 1000 dinucleosomes have been analyzed. Each State (#1 to #5) is illustrated by a typical AFM image of a dinucleosome in this state as defined on Figure 2, and the groups are defined as initial (I corresponding to State #1), intermediate (II corresponding to States #2 and #3) and final III (corresponding to States #4 and #5).