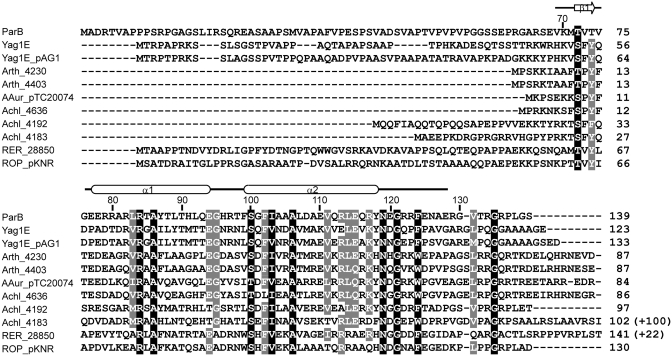
Figure 3.
Sequence alignment of ParB homologs. On the top, the residue number markers and secondary-structure elements observed in the crystal structure are indicated for ParB. The accession numbers of the 11 sequences in order are AAP69567 (ParB of pCXC100); ZP_03388950 (Yag1E in the chromosome of Propionibacterium acnes SK137); NP_052577 (Yag1E_pAG1 in plasmid pAG1 of Corynebacterium glutamicum); YP_829452 (Arth_4230 in plasmid 3 of Arthrobacter sp. FB24); YP_829155 (Arth_4403 in plasmid 1 of Arthrobacter sp. FB24); YP_950238 (AAur_pTC20074 in plasmid TC2 of A. aurescens TC1); YP_002478563 (Achl_4636 in plasmid pACHL02 of A. chlorophenolicus A6); YP_002477960 (Achl_4192 in plasmid pACHL01 of A. chlorophenolicus A6); YP_002477951 (Achl_4183 in plasmid pACHL01 of A. chlorophenolicus A6); YP_002766332 (RER_28850 in the chromosome of Rhodococcus erythropolis PR4); YP_002784447 (ROP_pKNR-00030 in plasmid pKNR of R. opacus B4). Sequence Achl_4183 is incomplete at its N-terminal region in GenBank and has been corrected here. The numbers in parentheses indicate extra C-terminal residues not shown. Residues with 100 and 80% conservation are shaded in black and gray, respectively.