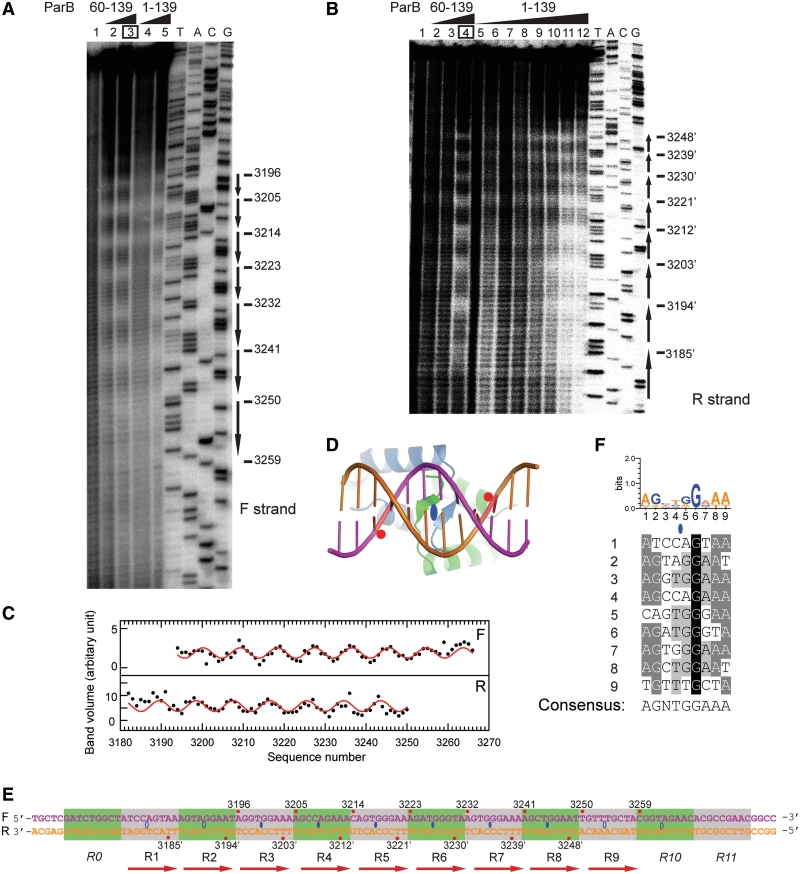
Figure 4.
The ParB-centromere interaction probed by hydroxyl radical footprinting. (A) Footprinting of the forward strand of ParB-bound centromere. A 170-bp centromere DNA was labeled at the 3′-end of the forward strand, assembled with 0.5 and 1 μM of ParB RHH domain (residues 60–139) (lanes 2 and 3), and 0.5 and 1 μM of full-length ParB (lanes 4 and 5) and subjected to hydroxyl radical cleavage. Lane 1 contains no protein. Sequencing ladders (A, T, G, C) are indicated. The most protected nucleotides derived from curve fitting are shown on the right. (B) Footprinting on the reverse strand. The ParB 60–139 concentrations were 0.25, 0.5 and 1 μM in lanes 2–4, and the ParB 1–139 concentrations were 0.1, 0.2, 0.25, 0.33, 0.5, 0.66, 1 and 2 μM in lanes 5–12. (C) The band volumes of the cleavage products are plotted against nucleotide numbers. Lane 3 (boxed) of the forward strand and lane 4 of the reverse strand were analyzed. Smaller volume corresponds to greater protection at the respective site. The lines represent the best fit of a sine function to the protection data with a period of 9.07 ± 0.08 for the forward strand (top panel) and 8.96 ± 0.09 for the reverse strand (bottom panel). (D) Structural model of the ParB RHH domain in complex with DNA. A standard B-form DNA was positioned based on the aligned pSK41 ParR-DNA complex structure (2Q2K). Red circles refer to the DNA backbone regions contacted by the N-terminus of helix α2. (E) The structure of the centromere. The forward strand is numbered from 5′ to 3′, and nucleotides in the reverse strand are numbered as their pairing nucleotides in the forward strand and are denoted by prime. Nucleotides with maximal protection in hydroxyl radical footprinting are marked by red circles and their nucleotide numbers. Ellipses denote binding centers of ParB dimer. Hollow ellipses are extrapolated from incomplete protection data. Alternative repeats are shaded in green and gray. Repeats R0, R10 and R11 do not bind ParB by themselves and are denoted in italics. (F) Alignment of repeats R1–R9 and the consensus sequence. The sequence logo is indicated at the top. A loose criterion of 50% conservation was used to define the consensus sequence.