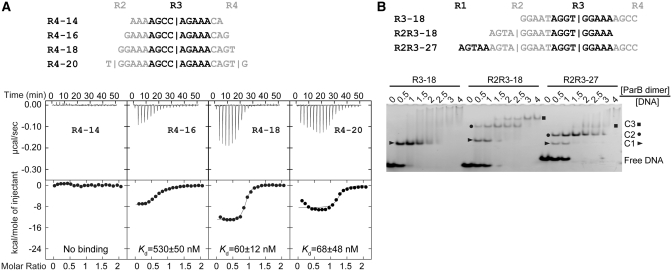
Figure 5.
The length requirement of single subsite for ParB binding and the binding stoichiometry. (A) ITC profiles of the ParB RHH domain (65–139) and the R4 probes of different lengths. The sequences of 14, 16, −18 and 20-bp R4 probes are indicated. Repeat R4 is shown in black and its flanking sequences are shown in gray. Vertical bars denote the predicted binding centers of ParB dimer. The curves are the best fit to a one-set-of-sites model. (B) Binding stoichiometry of ParB to one-site and two-site probes. Sequences are indicated for an 18-bp one-site probe R3–18; an 18-bp two-site probe R2R3–18; and a 27-bp two-site probe R2R3–27. Alternative repeats are shaded in black and gray. ParB 65–139 was assembled with 5 μM DNA in a 10 μl volume with different protein to DNA molar ratios as indicated. The native gel was stained by ethidium bromide. C1, C2 and C3 denote complexes formed by one, two and three ParB dimers, respectively.