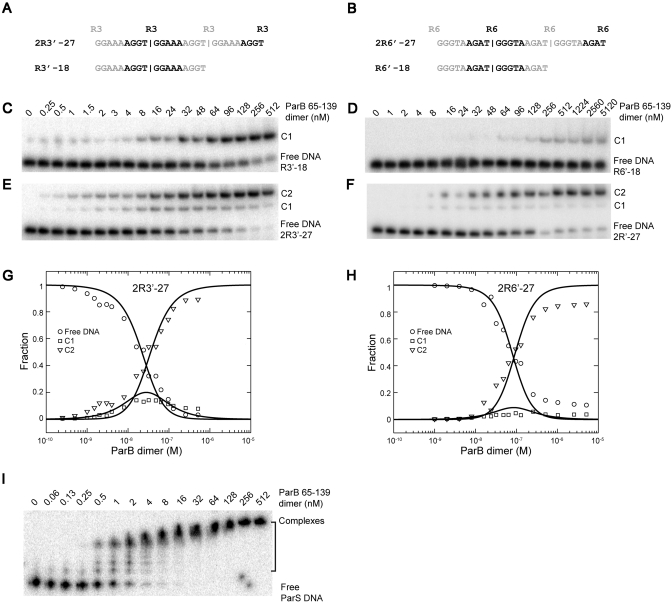
Figure 9.
The cooperativity of ParB binding to tandem identical repeats. (A and B) Sequence of two- and one-site probes designed for repeats R3 (A) and R6 (B). (C–F) EMSA of ParB 65–139 with one-site probe R3′–18 (C) and R6′–18 (D), and two-site probes 2R3′–27 (E) and 2R6′–27 (F). C1 and C2 denote complexes formed by one and two ParB dimers, respectively. (G and H) Global analysis of the binding isotherms for 2R3′–27 (G) and 2R6′–27 (H). The lines are the best fit to Equations (2–5), with intrinsic dissociation constant Kd,int = 112 nM and cooperativity ω = 16 for 2R3′–27; Kd,int = 877 nM and ω = 110 for 2R6′–27. (I) EMSA of ParB 65–139 with a 32P-labeled 170-bp DNA covering the whole centromere region.