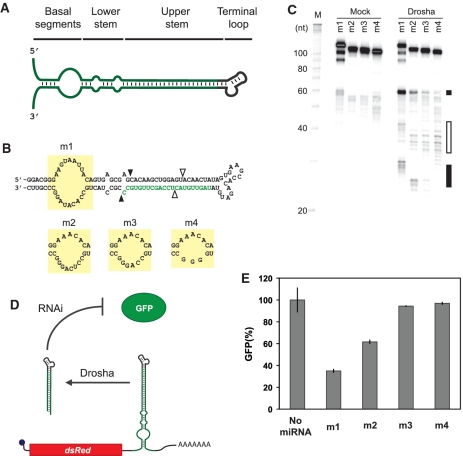
Figure 1.
Extent of structure in the basal segments dictates miRNA processing and target gene silencing. (A) General domains of a pri-miRNA. (B) Sequence and secondary structure of minimal pri-miRNAs with different bulge sizes in the basal segments. Mature miRNA sequence targeting GFP is shown in green. The bulge sequences are demarcated by yellow boxes. Black arrows indicate the predicted Drosha cleavage site for productive processing 11-bp downstream of the basal segments (23). White arrows indicate the predicted Drosha cleavage site for abortive processing 11-bp upstream of the terminal loop (23). (C) PAGE analysis of the minimal pri-miRNAs subjected to the Drosha cleavage assay. Black and white boxes mark the general locations of productive and abortive cleavage products, respectively. The three bands representing full-length m1 presumably arose from spontaneous cleavage of the unstructured bulge during or after gel extraction. (D) Schematic of the miRNA regulatory system in the cell-culture assay. GFP-targeting miRNAs were placed within the 3′ UTR of the transcript encoding DsRed-Express. 293 GFP cells transiently transfected with each construct were subjected to flow cytometry analysis. (E) Relative GFP levels obtained from the cell-culture assay for constructs harboring miRNAs with varying bulge sizes. See ‘Materials and Methods’ section for a description of data normalization. Error bars reflect the range of values from two independent transfections conducted in the same cell-culture plate. See Supplementary Figure S4A for results from an independent transfection experiment for (E).