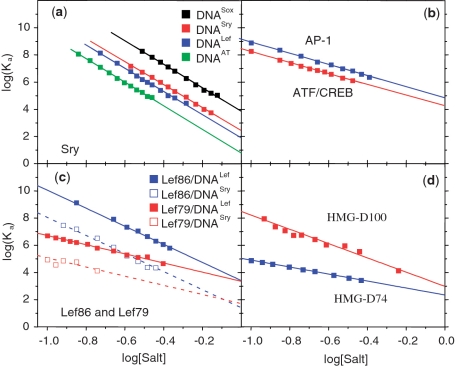
Figure 1.
The salt dependence of binding constants, Ka, (salt plots). (a) The SS HMG box from hSRY binding to its optimal target DNA (DNASox) and to three other duplexes of lower affinity, one containing just a central –ATAT– sequence (DNAAT). (b) The bZIP homodimer from yeast GCN4 binding to the AP1 (–ATGACTCAT–) and ATF/CREB (–ATGACGTCAT–) target sequences. (c) The HMG box from mLEF-1 (Lef86) and its truncated form lacking the basic C-terminal tail (Lef79) binding to DNALef (–TTCAAA–) the optimal target and to DNASry (–CACAAA–) a sub-optimal sequence. (d) The NSS HMG box from Drosophila HMG-D (D-100) and its truncated form (D74) lacking the basic C-terminal tail binding to DNALef. Data taken from Refs (11,12,17).