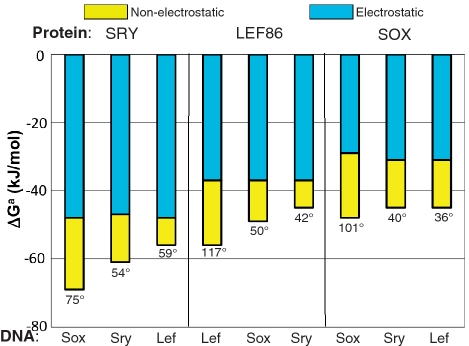
Figure 6.
Gibbs free energies of binding SS HMG boxes to their optimal and sub-optimal DNA sequences, separated into their electrostatic and non-electrostatic components using Equation (1). The protein-induced bend angles are given below the bars. Although the electrostatic component dominates the affinity in all cases, the more tightly a given HMG box binds (the more negative the total Gibbs energy), the greater is the non-electrostatic component and the larger the bend angle. As regards the relationship to the bend angle, see also (32). The DNA sequences DNALef, DNASry and DNASox are those previously considered optimal for binding the three HMG boxes, though SRY protein binds better to DNASox. Data taken from (12).