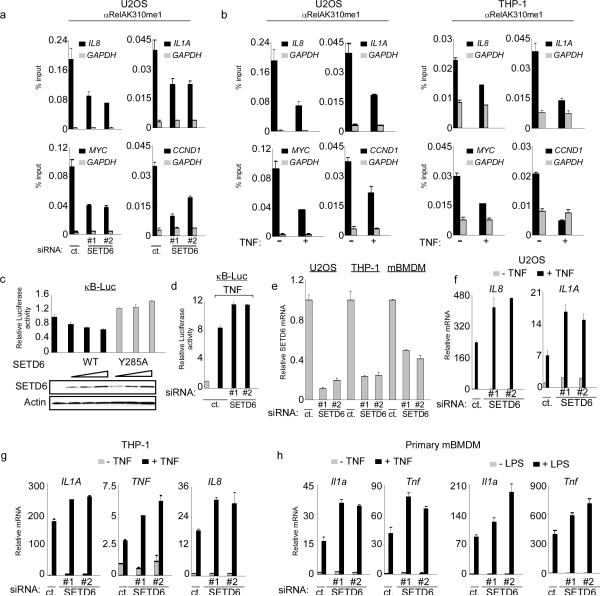
Figure 2. SETD6 monomethylation of RelA inhibits RelA transactivation activity.
(a) Enrichment of RelAK310me1 at promoters of RelA target genes required SETD6. Occupancy of RelAK310me1 in U2OS cells treated with control or SETD6 siRNAs at the promoters of IL8, IL1a, myc, ccnd1 (black bars) and gapdh (grey bars, as a control) was determined by real time (RT) PCR of ChIP samples. ChIP enrichment shown as % input (ChIP/input × 100). (b) Loss of RelAK310me1 occupancy at RelA target genes promoters in response to TNF treatment. ChIP assays as in (a) from U2OS (left) and THP-1 cells (right) ± TNF stimulation (20 ng/ml for 1hr). Negative control antibody ChIPs for (a) and (b) shown in Supplementary Fig. 9. (c) Repression of the NF-κB luciferase reporter (κB-Luc) by SETD6 was dose dependent and required an intact catalytic SET domain. κB-Luc reporter luciferase activity (normalized to Renila) and relative change compared to control transfection was determined 24 hrs after transfection of U2OS cells with increasing amounts of the indicated plasmids. Immunoblot analysis of SETD6 and SETD6Y285A expression is shown. (d) Depletion of SETD6 enhanced TNF-induced activity of an NF-κB driven reporter. Luciferase activity was determined as in (c) in 293T cells transfected with control or 2 independent SETD6 siRNAs ± TNF treatment (10 ng/ml for 1hr). (e) Efficiency of SETD6 mRNA knockdown by RNAi in the indicated cell lines was determined by RT PCR. (f–h) SETD6 depletion increased expression of RelA target genes in multiple cell types. RT PCR analysis of the indicated mRNAs from U2OS (f), THP-1 (g) and primary mouse BMDM (mBMDM) cells (h) transfected with control (ct.) or two independent SETD6 siRNAs ± TNF (20 ng/ml for 1 hr) or ± LPS (100 ng/ml for 1 hr) as indicated. Error bars in (a–h) indicate ± s.e.m. from at least three experiments.