Figure 3.
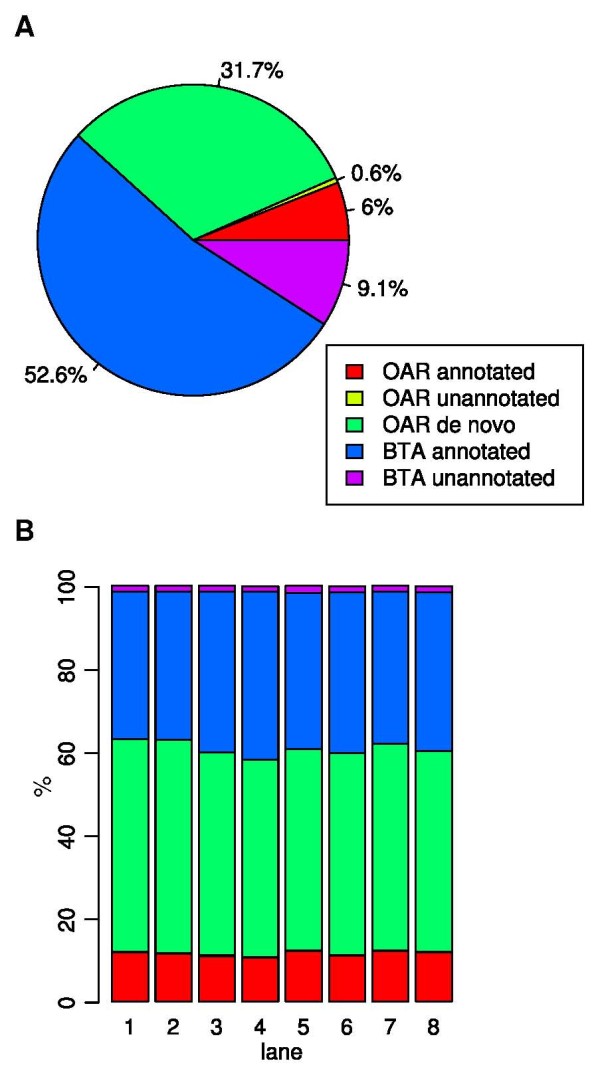
Composite reference transcriptome assembly and assignment of short read sequences. A De novo transcriptome assembly was performed with Velvet and Oases on the basis of mRNA sequence entries from GenBank for sheep and cow as well as the de novo assembled contigs from this study. All sequences were used as targets to map the short reads. The pie chart shows the relative proportions of the sequence entries from each of the sources used for the mapping (OAR = Ovis aries, BTA = Bos taurus). B The distribution of the targets that were matched by bowtie for short read mapping are shown. Most reads mapped to the de novo transcriptome assembly, but it was possible to map a substantial number of additional reads by use of the Ovis aries and Bos taurus mRNA sequences. Table 4 displays the exact counts for each lane.
