Abstract
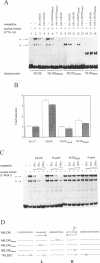
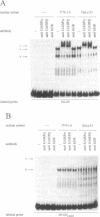
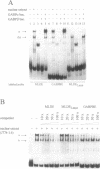
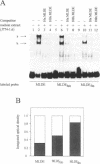
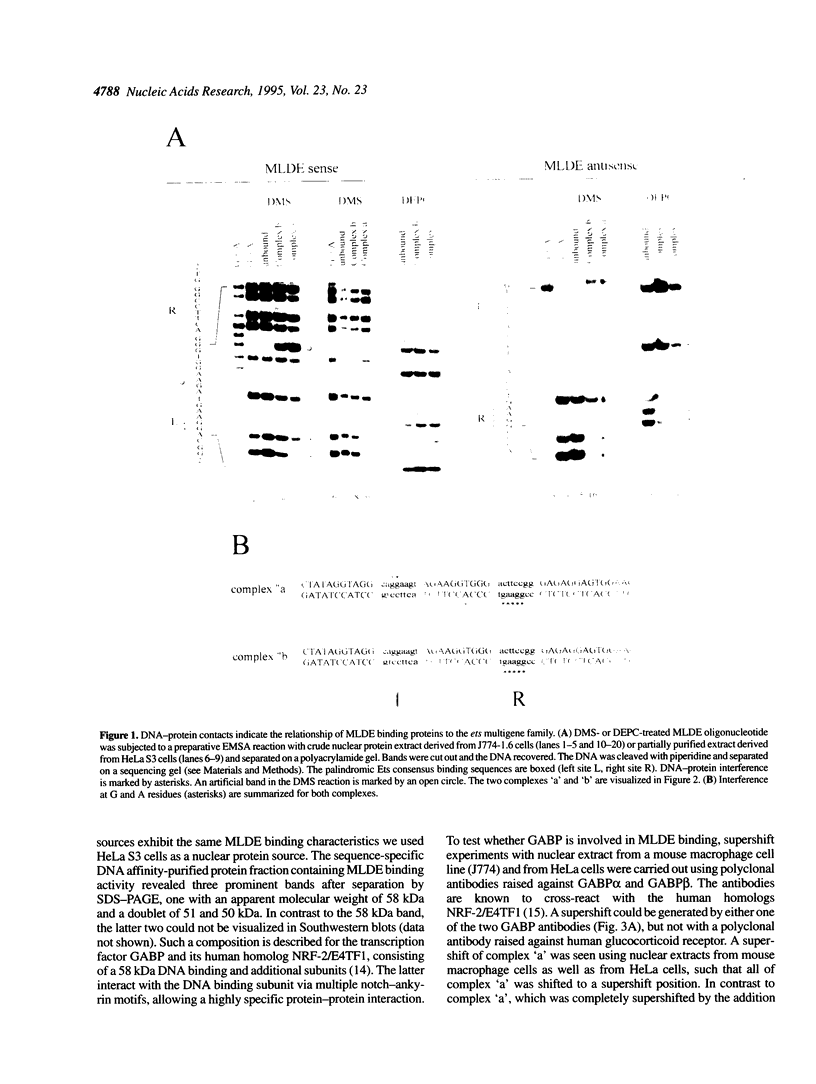
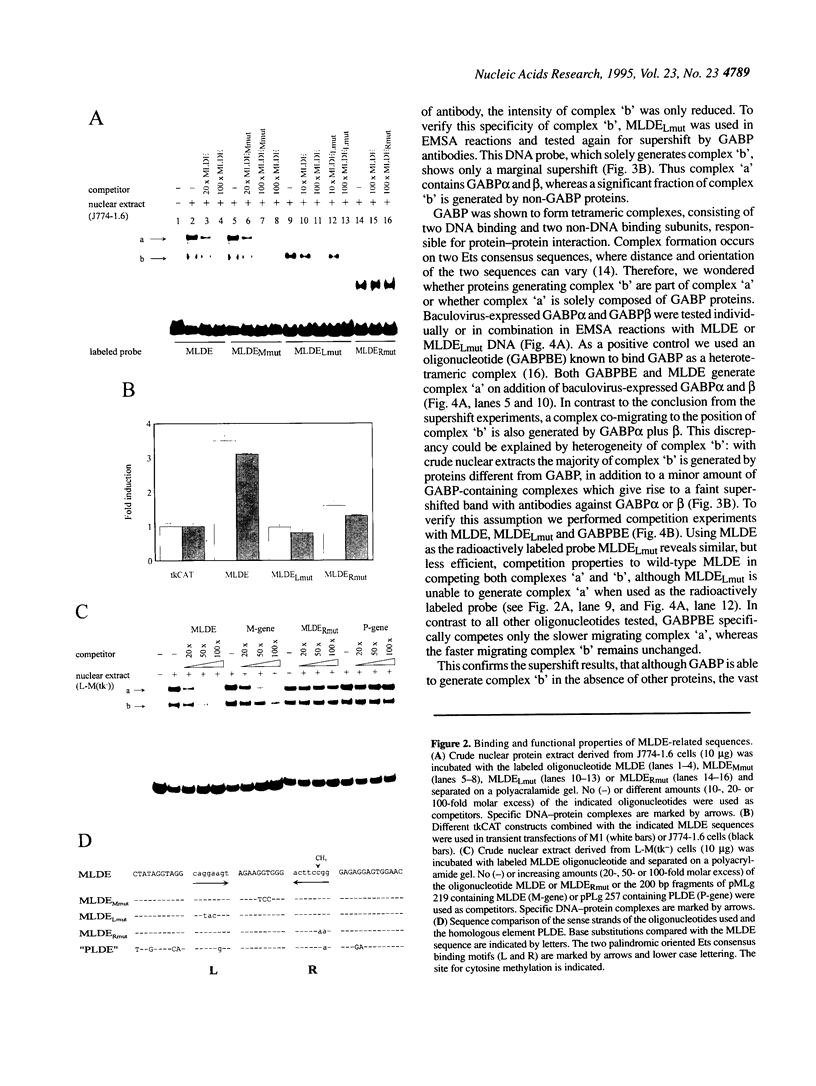
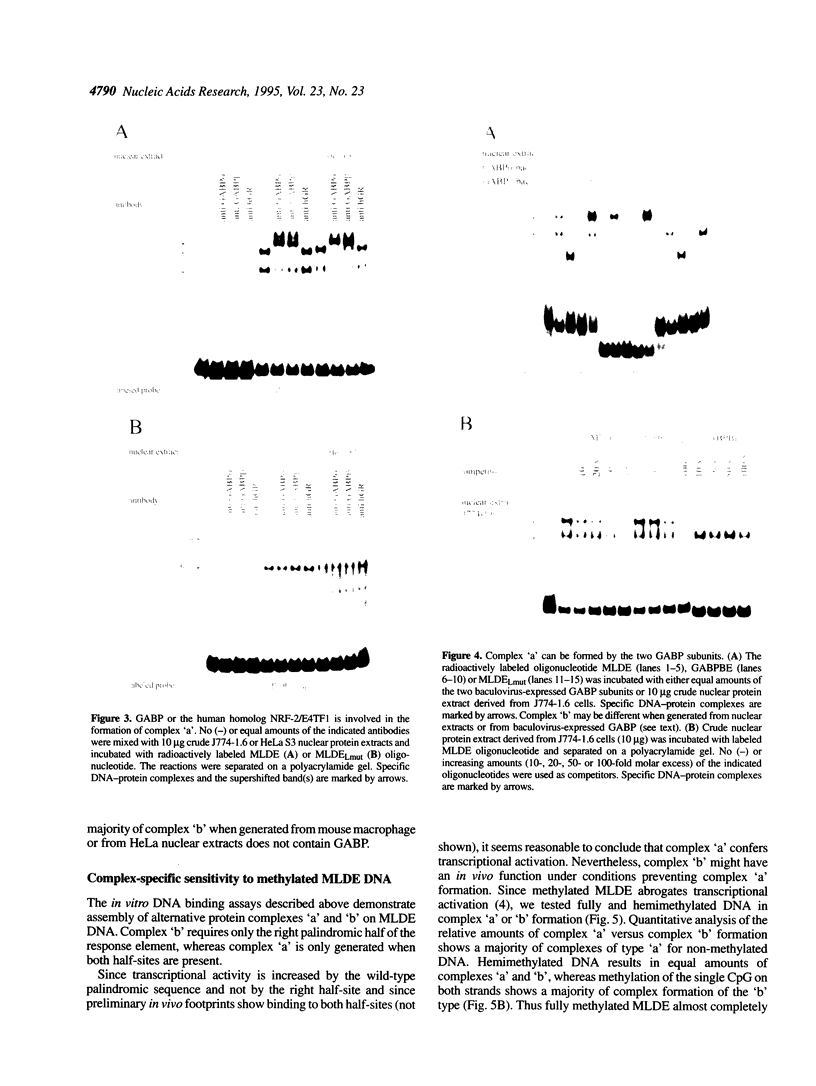
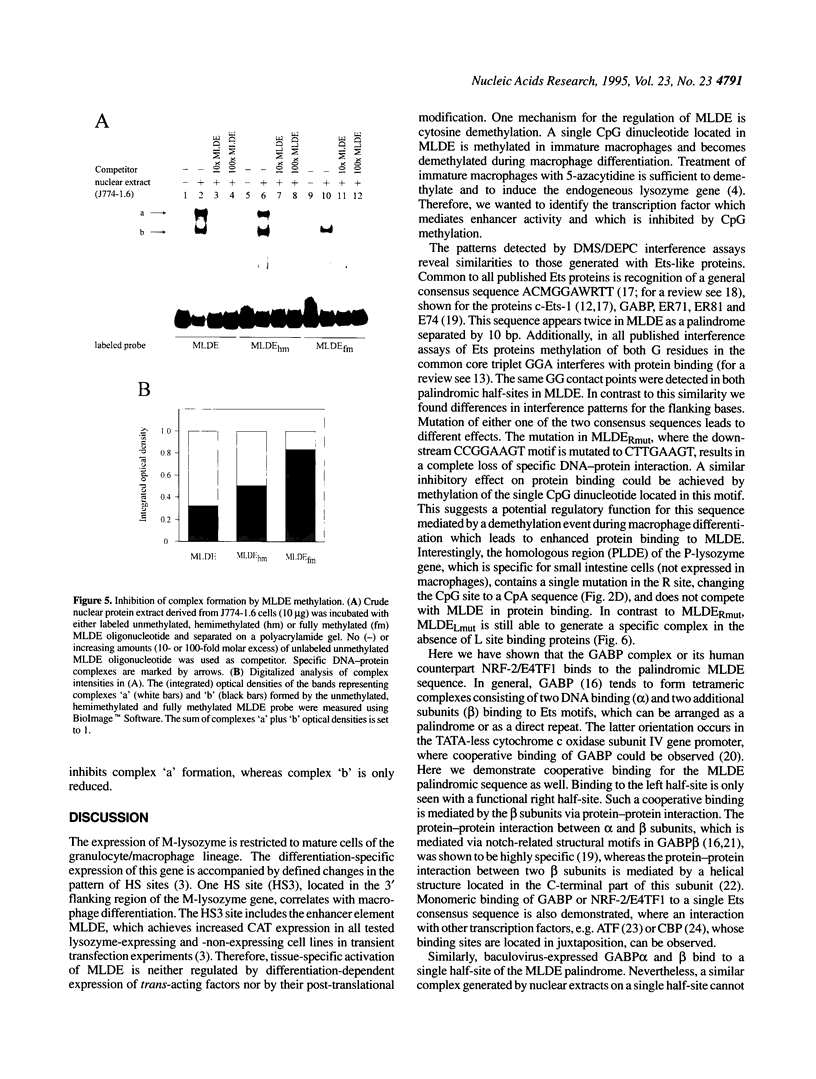
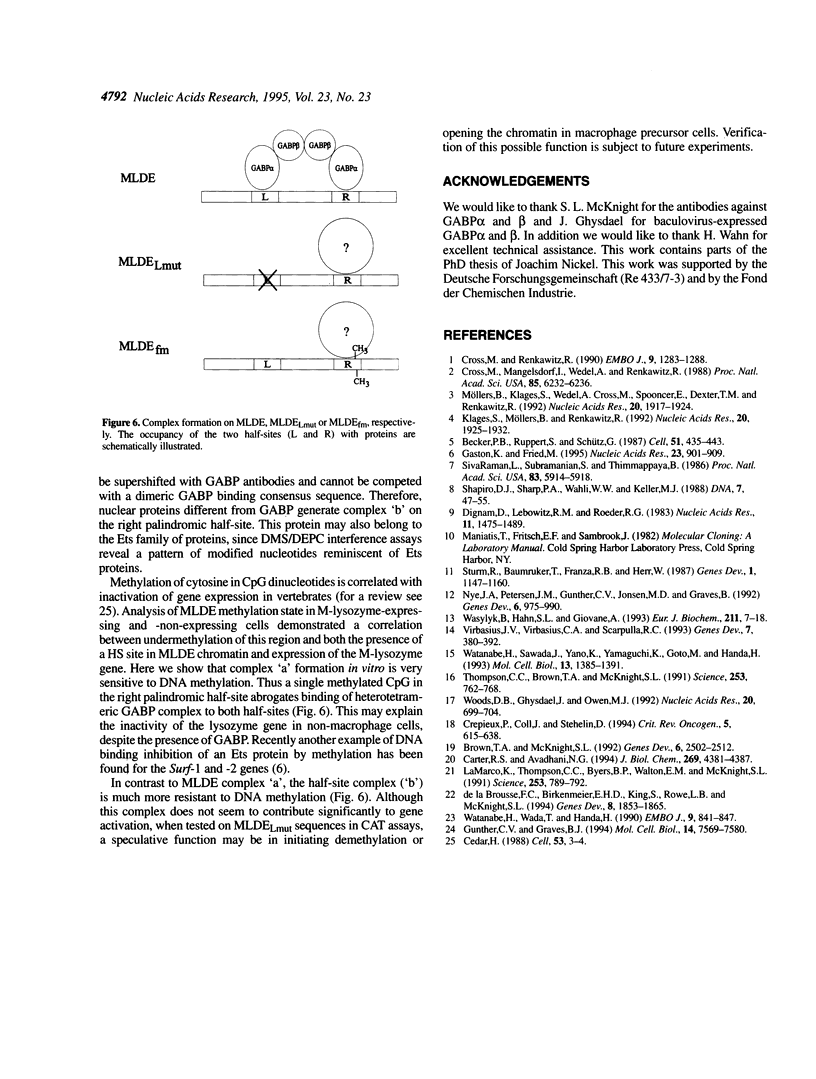
Expression of the mouse M-lysozyme gene is a specific marker for the differentiation of macrophage/granulocyte cell lineages. Analysis of the mechanisms regulating M-lysozyme gene expression revealed an enhancer element in the 3'-flanking region of the gene, termed the M-lysozyme downstream enhancer (MLDE). Here we demonstrate that the nuclear factors binding to MLDE are present in all tested myeloid and non-myeloid mouse cell lines. Sequence analysis of MLDE identified two different sequences, CAGGAAGT and CCGGAAGT, which match the consensus binding sequences for proteins of the ets gene superfamily. The two sites are oriented palindromicly and separated by 10 bp. DMS/DEPC interference assays revealed different patterns of DNA-protein contacts on the two sites. Mutation of each consensus sequence leads to an individual change in protein binding in vitro. Despite these differences, both sequences are bound by GABP, forming a heterotetrameric complex. Tissue specificity is correlated with demethylation of a single CpG dinucleotide located in one of the two Ets motifs. This site when methylated inhibits GABP binding to both sequences in non-macrophage cell types.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Brown T. A., McKnight S. L. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992 Dec;6(12B):2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- Carter R. S., Avadhani N. G. Cooperative binding of GA-binding protein transcription factors to duplicated transcription initiation region repeats of the cytochrome c oxidase subunit IV gene. J Biol Chem. 1994 Feb 11;269(6):4381–4387. [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Crepieux P., Coll J., Stehelin D. The Ets family of proteins: weak modulators of gene expression in quest for transcriptional partners. Crit Rev Oncog. 1994;5(6):615–638. [PubMed] [Google Scholar]
- Cross M., Mangelsdorf I., Wedel A., Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6232–6236. doi: 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M., Renkawitz R. Repetitive sequence involvement in the duplication and divergence of mouse lysozyme genes. EMBO J. 1990 Apr;9(4):1283–1288. doi: 10.1002/j.1460-2075.1990.tb08237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K., Fried M. CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 1995 Mar 25;23(6):901–909. doi: 10.1093/nar/23.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C. V., Graves B. J. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994 Nov;14(11):7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages S., Möllers B., Renkawitz R. The involvement of demethylation in the myeloid-specific function of the mouse M lysozyme gene downstream enhancer. Nucleic Acids Res. 1992 Apr 25;20(8):1925–1932. doi: 10.1093/nar/20.8.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarco K., Thompson C. C., Byers B. P., Walton E. M., McKnight S. L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991 Aug 16;253(5021):789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Möllers B., Klages S., Wedel A., Cross M., Spooncer E., Dexter T. M., Renkawitz R. The mouse M-lysozyme gene domain: identification of myeloid and differentiation specific DNasel hypersensitive sites and of a 3'-cis acting regulatory element. Nucleic Acids Res. 1992 Apr 25;20(8):1917–1924. doi: 10.1093/nar/20.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988 Jan-Feb;7(1):47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- SivaRaman L., Subramanian S., Thimmappaya B. Identification of a factor in HeLa cells specific for an upstream transcriptional control sequence of an EIA-inducible adenovirus promoter and its relative abundance in infected and uninfected cells. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5914–5918. doi: 10.1073/pnas.83.16.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm R., Baumruker T., Franza B. R., Jr, Herr W. A 100-kD HeLa cell octamer binding protein (OBP100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes Dev. 1987 Dec;1(10):1147–1160. doi: 10.1101/gad.1.10.1147. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Virbasius J. V., Virbasius C. A., Scarpulla R. C. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993 Mar;7(3):380–392. doi: 10.1101/gad.7.3.380. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993 Jan 15;211(1-2):7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Sawada J., Yano K., Yamaguchi K., Goto M., Handa H. cDNA cloning of transcription factor E4TF1 subunits with Ets and notch motifs. Mol Cell Biol. 1993 Mar;13(3):1385–1391. doi: 10.1128/mcb.13.3.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Wada T., Handa H. Transcription factor E4TF1 contains two subunits with different functions. EMBO J. 1990 Mar;9(3):841–847. doi: 10.1002/j.1460-2075.1990.tb08181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. B., Ghysdael J., Owen M. J. Identification of nucleotide preferences in DNA sequences recognised specifically by c-Ets-1 protein. Nucleic Acids Res. 1992 Feb 25;20(4):699–704. doi: 10.1093/nar/20.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Brousse F. C., Birkenmeier E. H., King D. S., Rowe L. B., McKnight S. L. Molecular and genetic characterization of GABP beta. Genes Dev. 1994 Aug 1;8(15):1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]