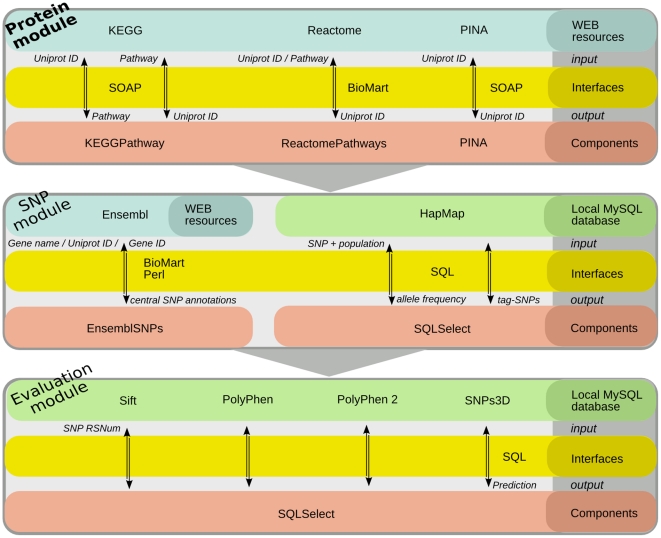
Figure 1. Workflow of CANGES modules and components, resources and interfaces within each module.
The input of the protein module is a list of protein identifiers or pathway names. The output is a list of corresponding focal genes, i.e., genes that belong to the same pathways with query protein or query pathway in KEGG or Reactome, or whose protein products interact with the query proteins. The resulted focal genes are inputs for searching the central SNPs (SNP module) using ENSEMBL and HapMap. The central SNPs are first queried from ENSEMBL and then the allele frequencies and tag-SNPs are fetched from a local HapMap database. The SNP module returns a list of central SNPs with their annotations and tag-SNPs. The list of central SNPs is then passed for the evaluation module in which the SNPs are then further evaluated with four methods that predict the functional effects of SNP to protein function.