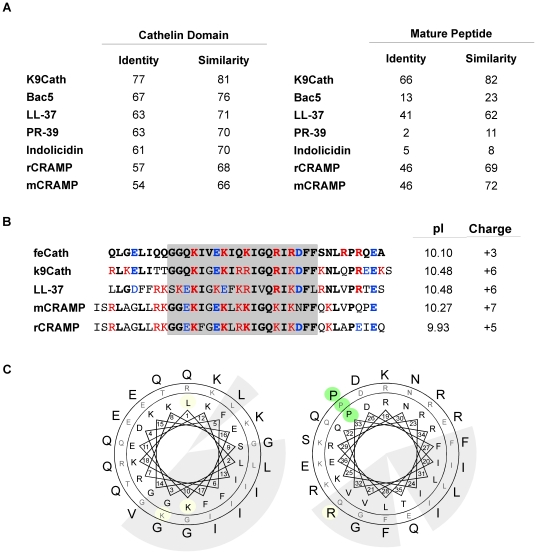
Figure 3. Amino acid comparison of cathelin-like domain and mature cathelicidin.
(A) Table of sequence identity and similarity of the cathelin-like domain from diverse groups of cathelicidins and the mature peptide of more closely related cathelicidins (PR-39 for distant comparison with mature peptides). K9CATH: dog, Bac5: cow, LL-37: human, PR-39: pig, Indolicidin: cow, mCRAMP: mouse, rCRAMP: rat. (B) Amino acid alignment of closely related mature cathelicidins. Residues in red are basic, residues in blue are acidic, and bolded residues are identical feCath sequence. pI and net charge are calculated for each peptide. Grey box indicates region of high similarity, and maintenance of cationic and hydrophobic residues. (C) Hydrophobic and polar residue clusters shown on a helical wheel projection. Based on the same analytical approach of Zelezetsky et al [25], sequence of feCath (outer ring) is compared to K9CATH (middle ring) and human LL-37 (inner ring). The analysis is divided into N- and C-terminal sequences, with residues 1–18 shown in the left wheel and 19–36 in the right wheel. Clusters of nonpolar residues are on a shaded background. Residues that deviate from the pattern of the three sequences are highlighted. The helical-breaking proline residue near the C-terminus is on a green background.