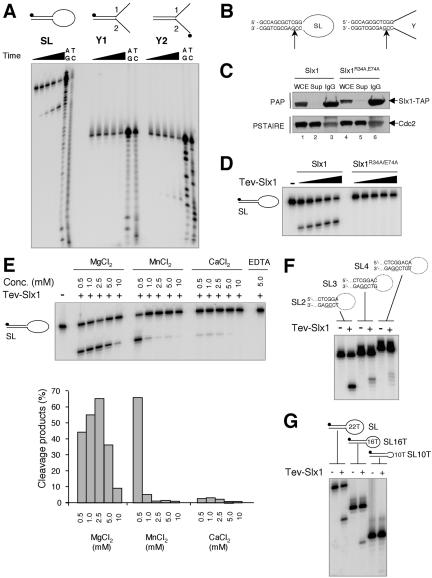
Figure 2.
Structure-specific DNA endonuclease activity associated with Slx1. (A) Three different substrates were incubated for 30 min at 30°C with 3 μl of TEV-Slx1 for 0 s, 30 s, 2.5 min, 10 min, and 50 min. A+G and C+T sequencing ladders were derived from the SL, y1, and y2 oligonucleotides. Reaction products were analyzed by denaturing PAGE. (B) The sites of cleavage were determined from the Maxam-Gilbert sequencing ladders and are indicated by arrows on each DNA structure. (C) Whole cell extract (WCE) derived from strains slx1::kanMx6 slx1wt-TAP:LEU1 and slx1::kanMx6 slx1R34A,E74A-TAP:LEU1 were incubated with IgG-Sepharose beads. TAP-Slx1 and TAP-Slx1R34A,E74A were detected in the WCE, supernatant (Sup), and the pulled down IgG-Sepharose beads (IgG) by Western blotting by using peroxidase-anti-peroxidase antibody (PAP; Sigma-Aldrich, St. Louis, MO). Cdc2 detected with an antibody directed against the Cdc2 PSTAIRE motif was used as a loading control. (D) SL was incubated with 1, 2, 4, 6, and 9 μl of TEV-eluate obtained from slx1/slx1wt-TAP::LEU1 and slx1/slx1R34A,E74A-TAP::LEU1 strains for 30 min at 30°C. Reaction products were analyzed by denaturing PAGE. (E) SL was incubated with 3 μl of TEV-Slx1 in the presence of various amounts of Mg2+, Mn2+, Ca2+, or EDTA and reaction products analyzed by denaturing PAGE. The relative amount of cleavage products was quantified by phosphorimager analysis. (F) The various SL substrates indicated above the gel were incubated with 3 μl of TEV-Slx1 for 30 min at 30°C. (G) Three microliters of TEV-Slx1 was incubated with SL containing ss loops of various sizes as indicated.