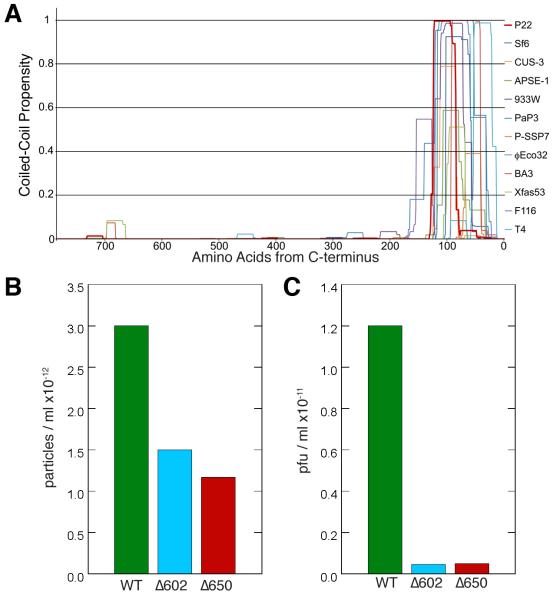
Figure 7.
(A) Plot of the conserved, predicted coiled-coil motif in the C-termini of portal proteins for several members of the Podoviridae family, as determined by the software COILS (Lupas et al., 1991) with a 21 amino acid scan window. The C-termini of the proteins are aligned at the right side of the plot. The phages encoding these portal proteins include: P22, Sf6, CUS-3 and APSE-1, divergent members of the P22-like phages; 933W, F116, BA3, Xfas53, P-SSP7, PaP3 and φEco32, podoviruses that are all distantly related to one another; and T4, a long-tailed myovirus. Each of these portal proteins shows a strong peak of coiled-coil probability within its C-terminal 150-100 a.a. For some, including P22 (in dark red), the probability drops in the terminal 70 a. a., even though the crystal structure clearly shows an uninterrupted coiled-coil extending up to within 6 a.a. of the C-terminus (Olia et al., 2011). The reason for this discrepancy between predicted and observed coiled-coil in P22 is not known. (B and C) C-terminal deletions of portal protein reduce phage assembly and infectivity. Phage particles/ml (B) and plaque forming units/ml (C) produced by trans-complementation of a P22 nonsense portal mutant with full length and portal proteins truncated from residues 603-725 (Δ602) and 651-725 (Δ650) as previously described (Moore and Prevelige, 2002). Fractions from a CsCl gradient that displayed peak infectivity were collected. The number of particles/ml was determined by DNA absorbance and the titer was determined on a permissive host. EM and gel analysis indicated that the particles produced with truncated portal were properly tailed and contained full length DNA as occurs in wild-type (WT) phage.