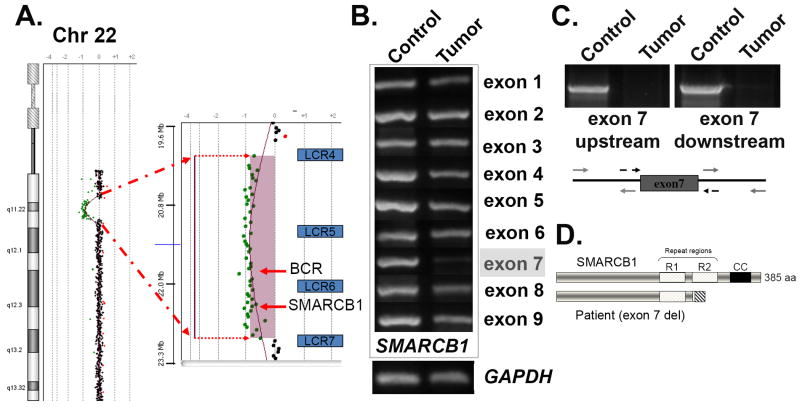
Figure 2. Germline mutation detection and molecular analysis of the rhabdoid tumor.
(A) A peripheral blood sample was used for comparative genomic hybridization (CGH) using the hg18 build on a U-array Cyto6000 platform. A ∼2.8Mb deletion resulted in partial monosomy (-1) in the chromosome 22q11.2 region, indicated by the green dots. A magnified view of the region shown on the right indicates loss of genetic material between the low copy repeat regions 4 and 7 (blu BCR and SMARCB1/hSNF5. (B) PCR analysis of the tumor sample. Chromosomal DNA was isolated following laser capture microdissection of rhabdoid tumor cells and used for PCR analysis. Each of the nine exons of SMARCB1 was individually amplified using both control and tumor cell DNA as templates. The GAPDH gene was used as a control for DNA concentration. Note the strongly reduced PCR product for exon 7. (C) PCR amplification of an extended region around exon 7 was used to verify that the entire exon was deleted in the tumor. The regions indicated by the grey arrows were used for amplification detection. (D) Schematic view of the deletion product that results in a frameshift and premature termination following exon 6. The SMARCB1 protein is shown (top), including the imperfect repeat regions (R1 and R2), as well as the C-terminal coiled-coil (CC). The deletion of exon 7 (lower diagram) results in the complete removal of residues within the R2 and CC regions are required for heterotypic protein interactions and assembly of SMARCB1 into SWI/SNF complexes.