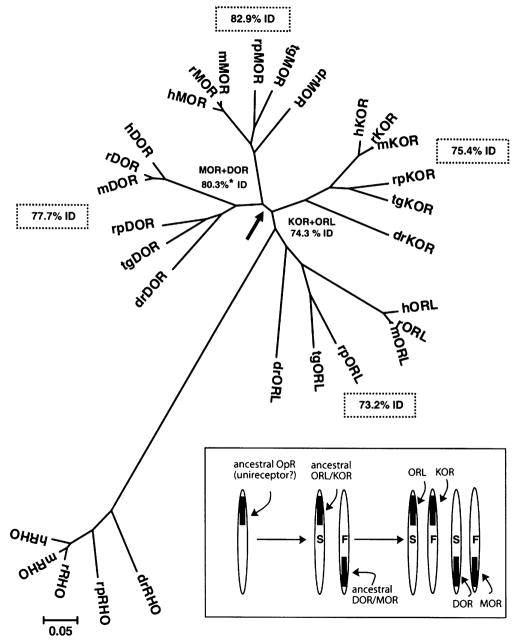
Fig. 2.
Phylogenetic analysis of MOR, DOR KOR and ORL sequences in six vertebrates. MEGA software was used to generate a radial phylogenetic tree using the neighbor-joining method, rooted with the available matching sequences of rhodopsin (RHO). Protein sequences from Rana pipiens (rp) were provided by conceptual translation of the cloned cDNA sequences deposited in GenBank. Abbreviations and access codes for MOR, DOR, KOR, and ORL sequences from other vertebrates were: dr: Dano rerio (zebrafish; AAK01143, AAP86771, AAG60607, AAN46747), tg: Taricha granulosa (newt; AAV28689, AAV28690, AAU15126, AAU26067), m: Mus musculus (mouse; P42866, P32300, P33534, P35377), r: Rattus norvegicus (rat; P33535, P33533, P34975, P35370), h: Homo sapiens (human; P35372, P41143, P41145, AAH38433). The arrow shows the bifurcation of MOR + DOR sequences from KOR + ORL. Values on plot are mean percent identity (%ID). Asterisk (*) indicates that these two values were significantly different by t-test. Branch length is equal to the proportional difference among the sequences (scale bar = 0.05 or 5% difference in amino acid sequence). Inset box: The molecular evolution of vertebrate opioid receptors. For simplicity, the genes are referred to by the same acronym as the opioid receptor proteins they encode. ‘S’ denotes slow and ‘F’ fast rate of adaptive evolution. See text for further details.