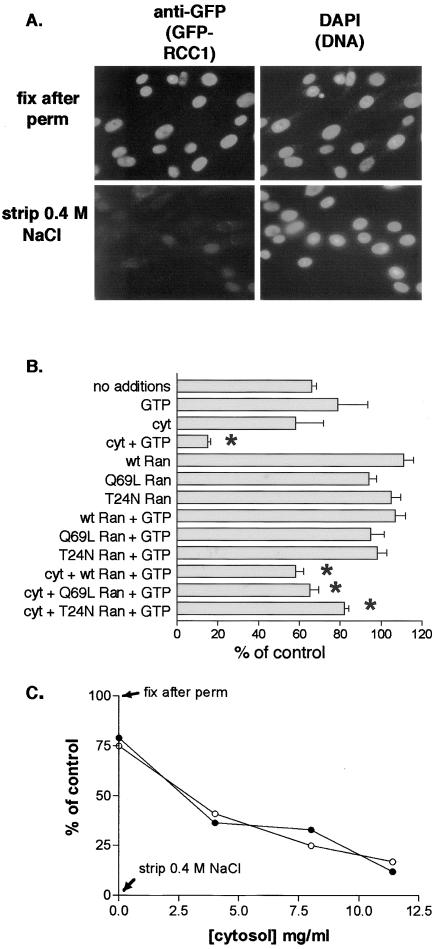
Figure 7.
Efficient GFP-RCC1 release from chromatin in permeabilized GFP-RCC1 tsBN2 cells requires cytosol and energy. (A) After permeabilization with 0.1% Triton X-100 and rinsing in buffer A, cells were either fixed immediately (top row) or incubated with buffer A containing 0.4 M NaCl for 20 min on ice before rinsing and fixation. GFP-RCC1 was localized with an anti-GFP first antibody and TRITC-labeled second antibody (left) and the cells were counterstained with 4,6-diamidino-2-phenylindole to localize the DNA. (B) After permeabilization, cells were incubated for 20 min at RT with the indicated additions before washing, fixation, and indirect immunofluorescence. GTP and Xenopus ovarian cytosol (cyt) were added at 1 mM and 11.5 mg/ml, respectively. wt, Q69L, or T24N Ran was added at 10 μM. Detection and quantitation of GFP-RCC1 was as described in MATERIALS AND METHODS. Shown for each condition are the mean and SEM of the nuclear fluorescence of between 30 and 90 nuclei. All values are expressed relative to the 100% (fixed after permeabilization) and the 0% (fixed after stripping with 0.4 M NaCl) controls. Note that the inclusion of 10 μM Ran decreased the loss obtained with cytosol alone (asterisks). (C) A concentration curve measuring the release of GFP-RCC1 obtained with increasing amounts of cytosol either in the presence (•) or absence (○) of 1 μM Ran. All samples contained 1 mM GTP.