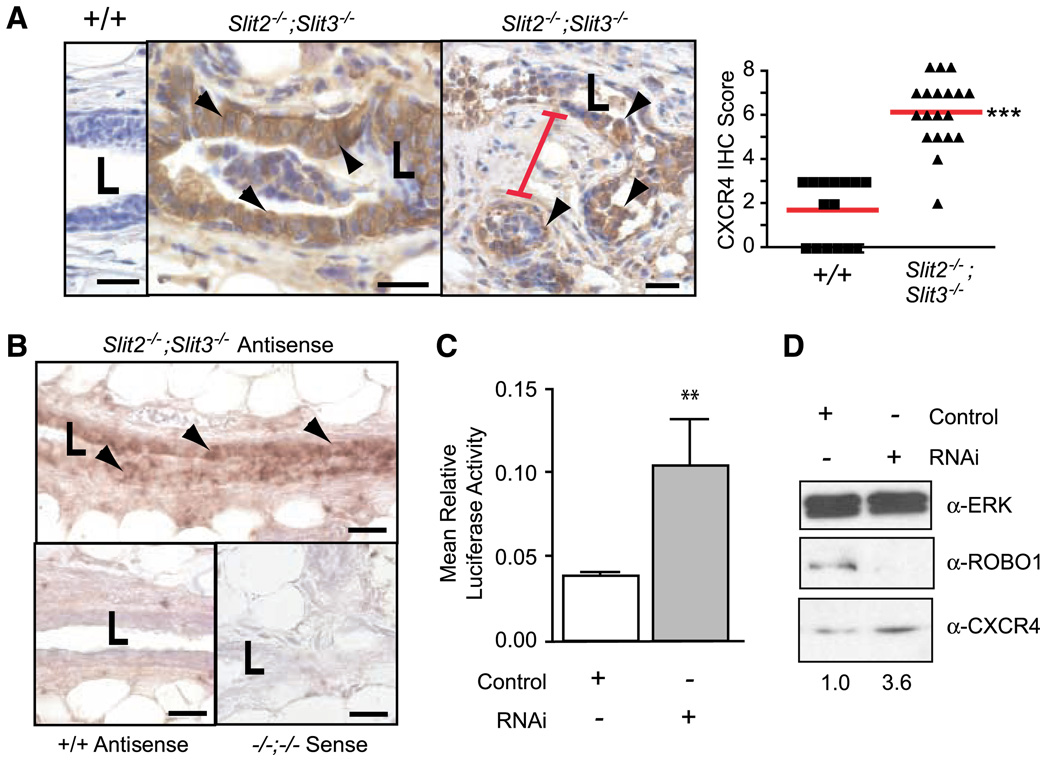
Figure 2.
Loss of Slit2 and Slit3 causes up-regulation of CXCR4 in mouse mammary gland and human MCF7 cells. A, in Slit2−/−;Slit3−/− outgrowths, CXCR4 protein expression is localized to epithelia, with desmoplastic stroma between lesions. Representative immunostaining with anti-CXCR4 on +/+ and Slit2−/−;Slit3−/− mammary outgrowths. Arrowheads, positive epithelial cells. Red bar, condensed desmoplastic stroma. Scale bar, 20 µm. CXCR4 immunostaining was scored according to positivity and staining intensity and plotted on a vertical scatter plot. Red bars, average score. Significantly more CXCR4 staining is seen in Slit2−/−;Slit3−/− outgrowths. ***, P < 0.0001, Mann-Whitney. B, Cxcr4 mRNA is specifically present in the epithelium of Slit2−/−;Slit3−/− outgrowths. In situ hybridization on +/+ and Slit2−/−;Slit3−/− outgrowths using antisense probes reveals Cxcr4 mRNA in Slit2−/−;Slit3−/−, but not +/+, cells. Arrowheads, positive epithelial cells. Sense probes show little or no background staining. Scale bar, 20 µm. L, lumen. C, loss of SLIT/ROBO signaling in MCF7 cells leads to up-regulation of Cxcr4 gene expression. Cells were treated with control or Robo1 siRNA and then cotransfected with pGL-CXCR4(−375), which contains the Cxcr4 promoter region coupled to the F-luciferase gene and pRL-TK (R-luciferase). Cells were lysed after 36 h and luciferase activity was measured in triplicate. Activities were normalized for transfection efficiency. Columns, mean relative luciferase activity; bars, SE. **, P = 0.0095, Mann-Whitney test. D, loss of SLIT/ROBO signaling in MCF7 cells leads to increased levels of CXCR4 protein. Representative immunoblots (n = 4). Numbers, CXCR4 band intensity.