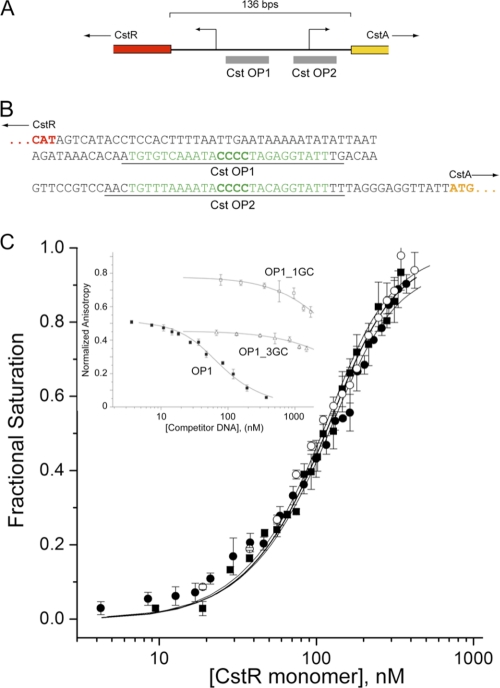
FIGURE 4.
Reduced CstR binds to cst operator (CstO) sites with high affinity in a manner dependent of the central run of four GC base pairs. A, schematic of the cstR-cstA intergenic region highlighting the positions of the two tandem candidate CstR operator sites. B, nucleotide sequence of the cstR-cstA intergenic region, highlighting the OP1 and OP2 operator sequences (green). Underlined bases correspond to the 5′-fluorescein-labeled duplex oligonucleotides used for DNA binding experiments. C, CstR binding isotherms for OP1 (solid squares, ■), OP2 (open circles, ○), and OP1_5GC (filled circles, ●) in which an additional GC base pair was inserted into the run of four GC base pairs. Inset, competition dissociation experiments with fluorescein-labeled apo-CstR-OP1 complexes with unlabeled wild-type OP1 (solid squares, ■), OP1-GC3 (open triangles, △), and OP1_1GC (open squares, □) duplexes. Fitted parameters derived from a two-tetramer binding model (15, 16) are compiled in Table 1. Conditions used were pH 7.0, 0.2 m NaCl, at 25.0 °C.