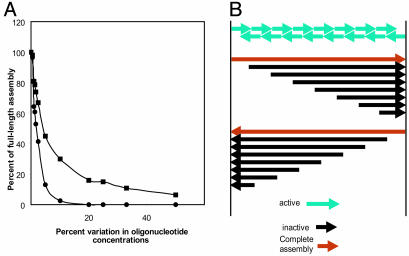
Fig. 2.
(A) Computer simulation of the ligation reaction as a function of variation in the concentrations of the input oligonucleotides. Computer simulation parameters are 130 top oligonucleotides (with bottom oligonucleotides saturating) and 2,000 molecules of each top oligonucleotide. Percent variation of oligonucleotide concentrations is determined by the randomly chosen number of molecules of each oligonucleotide. For example, at 10% variation in oligonucleotide concentrations, the number of each oligonucleotide is randomly chosen between 1,800 and 2,000. At each iteration of the program, a random pair of assemblies is selected and the pairs are joined together to form a larger assembly if the end coordinate of one assembly is one less than the beginning of the other assembly. The process is iterated until the number of assemblies no longer changes during 1 million pairings. -▪-▪-, average percent length of assemblies; -•-•-, percent of full-length assemblies. (B) The final products of a theoretical PCA reaction that starts with oligonucleotides of uniform size. Only the two terminal oligonucleotides can be extended to full length because polymerization is only in the 5′ to 3′ direction. An assembly is considered “active” if it can be extended by overlapping with another assembly followed by fill-in synthesis. An assembly is “inactive” if it cannot be extended further. Mass increase factor = final mass/beginning mass = sn (n + 2)/2sn = (n + 2)/2 ≈ n/2. Fraction of final mass full length = [2 (sn + s/2)]/[sn (n + 2)] = [2 (n + 1/2)]/[n (n + 2)]≈ 2/n. s = nucleotide length of oligonucleotide; n = number of oligonucleotides on one strand.