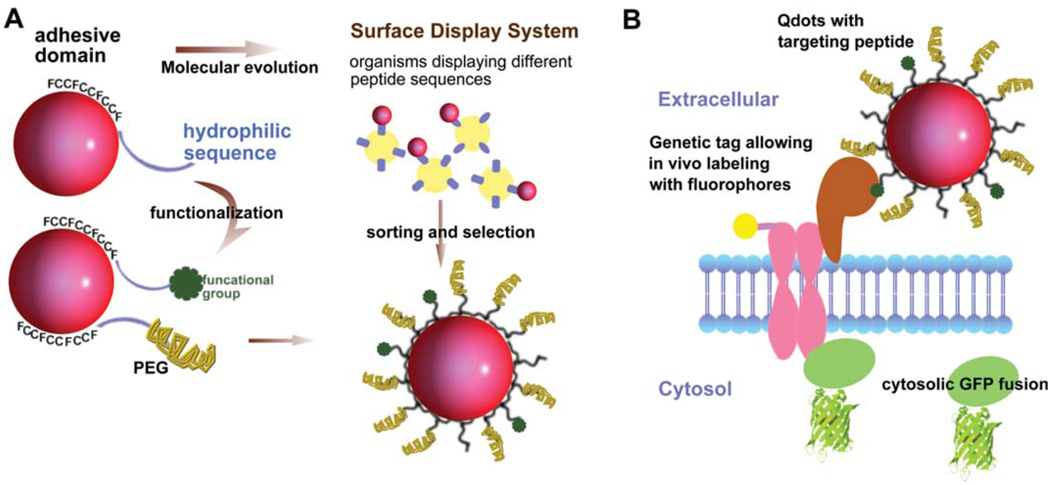
Figure 2.
(online colour at: www.biophotonics-journal.org) (A) Design and evolution of peptide-coated Qdots. The natural peptide used for Qdots solubilization has a hydrophobic adhesive domain and a hydrophilic sequence [44, 48]. The hydrophilic tail can be functionalized by adding reactive groups or functional groups such as biotin at the N-terminus. Polyethylene glycol (PEG)-conjugated peptide can be introduced simultaneously to reduce non-specific binding to cell surfaces. The adhesive domain sequence can be selected via peptide display libraries such as phage or yeast surface display system. This method can be used to select for peptides that specifically bind to Qdots, or even peptides that improve the photophysical properties such as reduced blinking or enhanced brightness. (B) Schematic representation of multiplexing fluorescent labeling methods to study protein recruitment, trafficking, and activation. Qdots (in red) with specific targeting peptides, genetic tags allowing organic fluorophores to be covalently attached (fluorophore in yellow), and FP tags on cytosolic proteins (in green) can be used in parallel to study complex, dynamic system such as signal transduction pathway.