Figure 1.
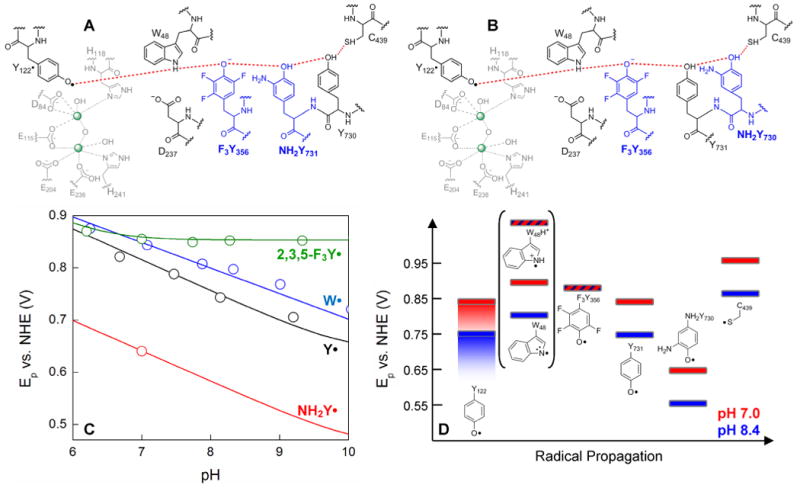
The proposed radical initiation pathway and its energetics in E. coli RNR with site-specific incorporation of unnatural amino acids (13). Residues in grey are associated with the diferric cluster, in black are proposed pathway residues (13), and in blue are the unnatural amino acids (F3Y and NH2Y) to probe the pathway in the present studies. Note that the structural location of Y356 is unknown. (C) Peak potentials (Ep) for free (NH2Y), and N-acetylated and C-amidated (Y, W, F3Y) amino acids, as a function of pH. The Eps for Y•, W• and 2,3,5-F3Y• have been previously determined (25) and the trace for NH2Y• has been generated from the reduction potential determined at pH 7, assuming Nernstian behavior (27). (D) The Eps from panel (C) have been assigned to residues in the radical propagation pathway to provide a qualitative energy landscape. Red and blue rectangles represent the peak potentials for each amino acid at pH 7.0 and 8.4, respectively. The peak potentials of WH•+ and F3Y• are represented by red rectangles with blue diagonal lines as they do not change between pH 7–8.4. The Ep range for Y122• is expanded (indicated by shading), because its properties relative to the other three Ys, including its pKa, are unique. Y122• has a half-life of ∼4 days and likely represents a thermodynamic hole (45). Brackets are placed around W48 as no direct evidence is available that places it on the pathway. If it is on the pathway, its protonation state, W48H•+ vs. W48•, that participates in radical transfer is unknown, and therefore Eps for both of these species are included.
