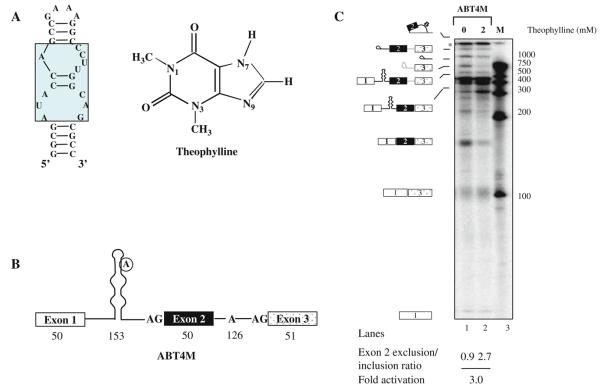
Fig. 10.2. Ligand-dependent modulation of alternative splicing using theophylline-binding aptamer in vitro.
(A) Diagram of the theophylline RNA aptamer sequence (left panel) and the chemical structure of theophylline (right panel ). The residues that are conserved for theophylline binding are enclosed in the rectangular box. (B) Schematic representation of a theophylline-binding aptamer sequestering the branchpoint of a model pre-mRNA. The encircled adenosine residue represents branch nucleotide. Open boxes represent exon sequences and horizontal lines between exons indicate introns. The numbers indicate size of the exon or intron. (C) In vitro splicing of ABT4M pre-mRNA. 32P-labeled ABT4M pre-mRNA was subjected to in vitro splicing in the absence (lane 1) or presence of 2 mM theophylline (lane 2). The extracted RNAs were fractionated on a 13% polyacrylamide denaturing gel. Schematic representations of the various RNA species are indicated on the left. Molecular Dynamics PhosphorImager and the ImageJ software were used to quantitate the products of the in vitro and in vivo RNA splicing reactions (see Fig. 10.3).