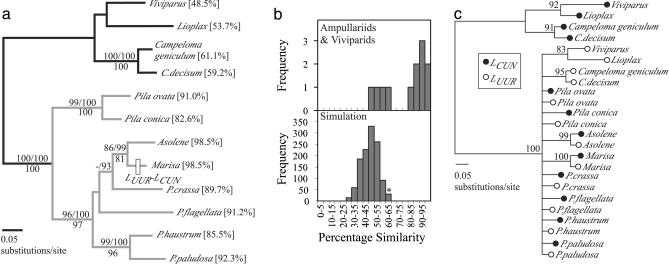
Fig. 1.
(a) ML phylogeny of eight ampullariid gastropods (genera Pila, Pomacea, Marisa, and Asolene) and four viviparid outgroups (genera Viviparus, Lioplax, and Campeloma) based on 678 bp of mtDNA. Support (>70%) for each clade is shown above branches for ML (100 replicates) and MB (posterior probabilities) and below for MP (1,000 replicates). ML, MB, and MP consensus trees were topologically congruent except for one clade: Parsimony analyses weakly supported a clade of Pomacea crassa and Pomacea flagellata (support: 60%) that was not recovered under ML and MB. A derived gene order change was discovered in Marisa only. Percent similarity values for LCUN vs. LUUR comparisons are given next to taxon labels; a clade with similarity values >75% is shown in gray. (b) The distribution of leucine tRNA similarity values sampled for the 12 ampullariids and viviparids (Upper) versus the simulated evolution of leucine tRNAs along the ML phylogeny (Lower). The asterisk refers to the starting value in our simulation model (61%). (c) A midpoint rooted 50% majority rule consensus phylogeny (MB) of 12 LCUN and 12 LUUR sequences from eight ampullariid taxa and four viviparid outgroups. Posterior probabilities (>70%) are shown above each node.