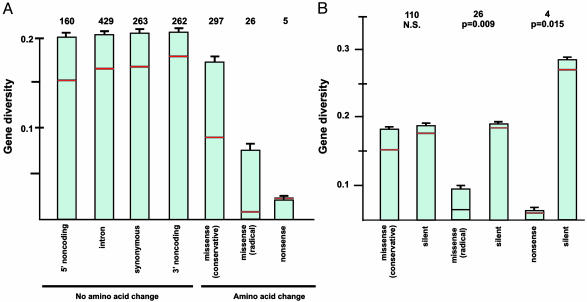
Fig. 1.
(A) Mean gene diversity (heterozygosity) at SNP sites categorized by location in the gene and effect on protein coding. Numbers of SNP sites in each category are indicated. Error bars indicate standard errors and red horizontal lines indicate median values. One-way analysis of variance, F6,1435 = 3.77; P = 0.001. All means except that for nonsense SNPs were significantly different from that for missense, radical SNPs (Dunnett's test; family error rate of 0.05 and individual error rate of 0.0167). Medians were significantly different by the Kruskal–Wallis nonparametric analysis of variance, P = 0.001. (B) Mean gene diversity compared pairwise between replacement (conservative missense, radical missense, and nonsense) SNP sites and silent SNP sites in the same genes. Error bars indicate standard errors and red horizontal lines indicate median values. Numbers indicate numbers of loci. Significance levels for paired-sample t tests are shown. Significance levels for Wilcoxon paired tests of the equality of medians were as follows: conservative missense, n.s.; radical missense, P = 0.017; nonsense, n.s.)