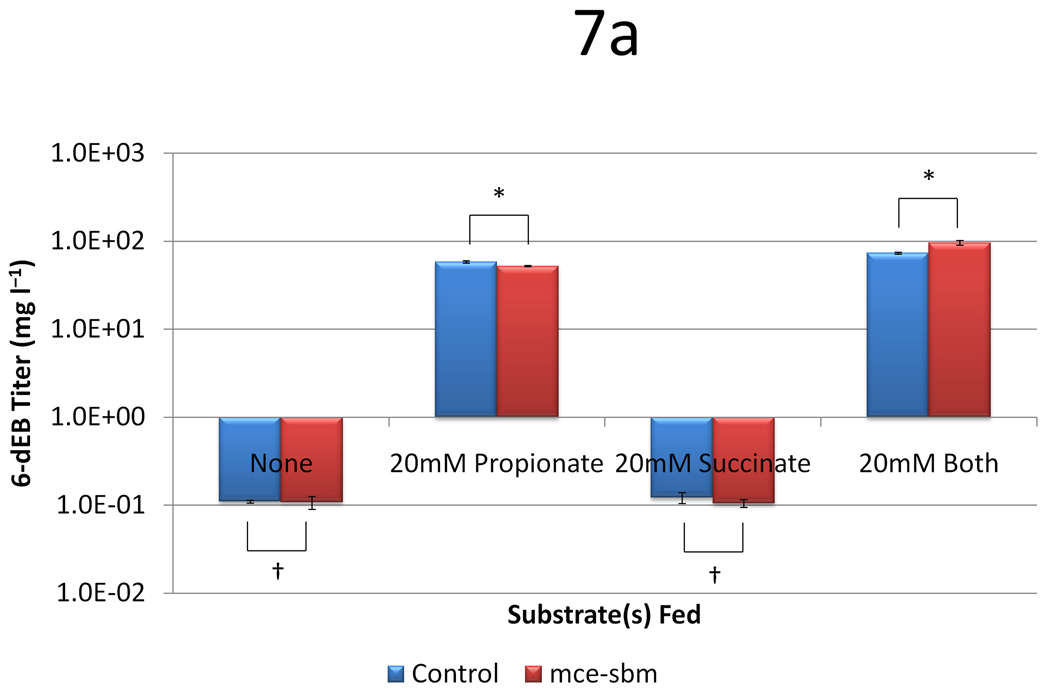
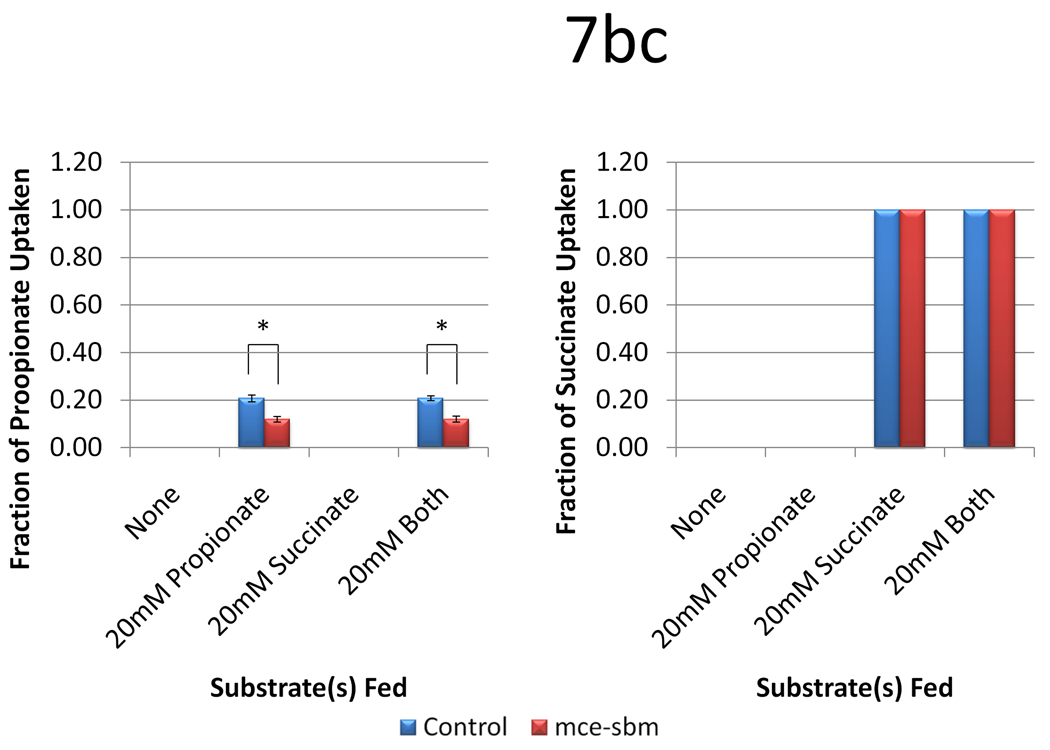
Figure 7.
Data from engineering the methylmalonyl-CoA mutase-epimerase pathway as a function of two plasmid systems (a control with only pBP130 and pBP144, and pCDFDuet-mce-sbm) and four medium formulations (no substrates, 20 mM propionate, 20 mM succinate, or 20 mM both substrates). Panel (a) shows the 6-dEB titer, panel (b) shows the fraction of propionate utilized, and panel (c) shows the fraction of succinate utilized as a function of these cellular parameters. Error bars represent ± one standard deviation from three replicates. * indicates statistically significant results (p < 0.05), while † indicates statistically insignificant results (p > 0.05) between the comparisons shown.