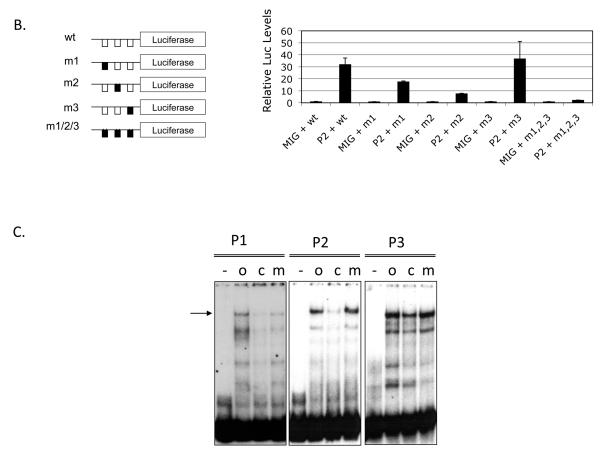
Figure 2. Mpl expression is upregulated by PLAGL2 in hematopoietic cells.
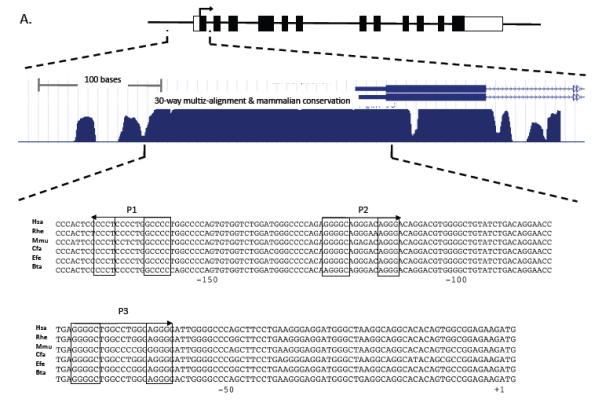
(A) Top: depiction of the Mpl gene structure, including untranslated exon sequences (blank boxes), coding regions (black boxes), translation start (arrow). Middle: zoom-in of the proximal promoter, exon 1 (black box) and section of intron 1 (arrowed line), and 30-way multiz-alignment & mammalian conservation analysis of the mouse genome segment chr4:118,129,933-118,130,333; using the UCSC Genome Browser on Mouse July 2007 (NCBI37/mm9) Assembly. Bottom: ClustalW sequence alignment of the Mpl proximal promoter (−189 to +3 from translation start site) including human (Hsa), monkeys (Rhe), mouse (Mmu), dog (Cfa), horse (Efe), and cow (Bta). The PLAGL2 consensus binding sites GRGGC(6-8)RGGK (P1 and P2) and a third site (P3) with an (N)9 linker are shown in boxes. Arrows indicate orientation of binding site. (B) Analysis of PLAGL2 activation of the Mpl proximal promoter using luciferase reporters in NIH3T3 cells. The luciferase reporters with 300 bp of the Mpl proximal promoter (wt) and respective mutants ablating one or more sites (m1, m2, m3, m1, 2, 3) are illustrated on the left panel. Fold activation relative to wt reporter and no PLAGL2 (MIG + wt) of each construct with (PL2) or without (MIG) PLAGL2 is shown on right panel. (C) Electrophoretic mobility shift assay of PLAGL2 consensus sites P1, P2, and P3 in the Mpl proximal promoter, using labeled wild type oligo (o), unlabeled oligo (c), or labeled oligo with point mutation in the core box of the Consensus PLAGL2 site (m). Arrow indicates PLAGL2:DNA binding.