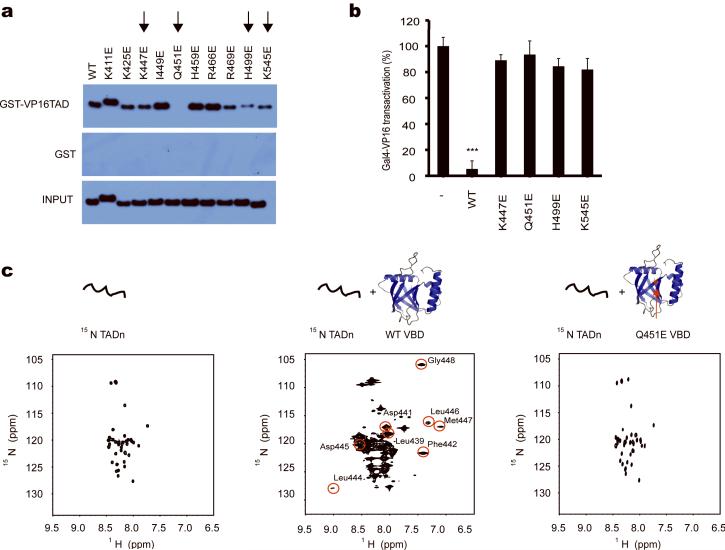
Figure 5.
Functional studies of mutant MED25 VBD. (a) VP16 full-length TAD-pull-down assays with MED25 VBD mutants. K447E, Q451E, H499E and K545E-mutated MED25 VBD exhibit weak or no binding to VP16 TAD (marked with an arrow). (b) While wild-type MED25 VBD acts in a dominant negative fashion to inhibit Gal4p-VP16 full-lenth TAD-dependent transcription, the K447E, Q451E, H499E and K545E-mutated MED25 VBD are incapable of inhibiting VP16-mediated transcription (***: p<0.01, t-test, one tailed, error bars represent s.d.). (c) The MED25 VBD Q451E mutation on β3 adjacent to the hydrophobic pocket disrupts binding of VP16 TADn to MED25 VBD. 1H-15N-HSQC spectra of free VP16 TADn (left panel), at 1:1.3 excess of wild-type MED25 VBD (middle panel) and with 1:1.5 excess of Q451E MED25 VBD (right panel) show that VP16 TADn only loosely binds, as seen by minor chemical shift changes, to mutant MED25 VBD without adopting a folded conformation. All far-shifted signals caused by the addition of wild-type MED25 VBD (middle panel, circled in red with assignment indicated) are missing when the mutant MED25 VBD is added to 15N-labeled VP16 TADn (right panel).