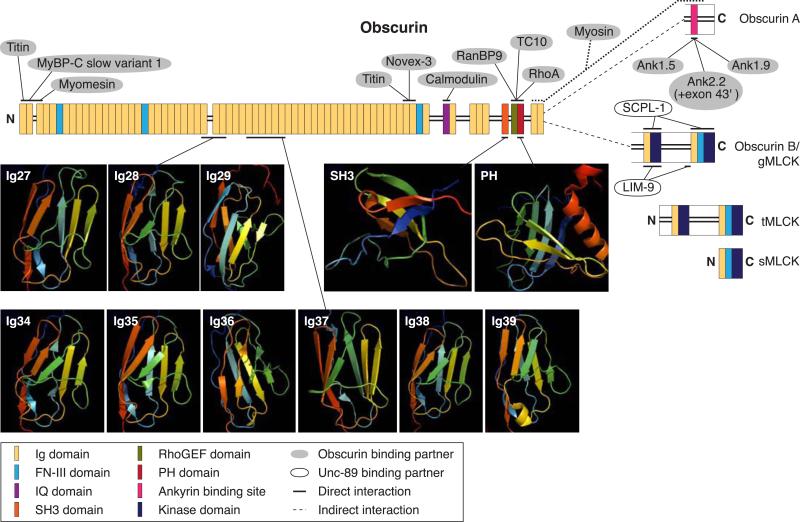
FIG. 5.
Schematic representation of the known isoforms of obscurin, illustrating their motifs, structures, and binding partners. Alternative splicing of the OBSCN gene results in at least four forms of obscurin: two giant isoforms, Obscurin A and Obscurin B/giant (g) MLCK, and two smaller kinase containing isoforms, Obscurin tandem (t) MLCK and Obscurin single (s) MLCK. All isoforms are composed of multiple structural motifs and signaling domains (see key for complete domain list and color coding). The giant isoforms diverge at the COOH terminus. Obscurin A is composed of a nonmodular COOH terminus, which houses the ankyrin-binding site (maroon box). The COOH terminus of Obscurin B/gMLCK possesses two kinase domains along with two additional Ig domains and a FN-III domain. Only 11 of obscurin's many domains have been characterized structurally, either by NMR or X-ray crystallography, which are shown as a cartoon model with flat arrows and coils representing β-sheets and α-helices, respectively. Binding partners and interaction sites that have been mapped to obscurin are also shown. Binding partners shown to interact directly with obscurin are depicted with a gray background and solid line pointing to their binding site on obscurin. The interaction between obscurin and myosin is not as well defined and may only be indirect and is thus shown by a dotted line. The phosphatase SCPL-1 and the LIM-domain protein LIM-9 were shown to interact directly with the kinases and preceding Ig and FN-III domains of Unc89, obscurin's invertebrate homolog, and are shown with a white background.