Abstract
The regulation of RNA synthesis by RNA polymerase (RNAP) is essential for proper gene expression. Crystal structures of RNAP reveal two channels: the main channel that contains the downstream DNA and a secondary channel that leads directly to the catalytic site. Although nucleoside triphosphates (NTPs) have been seen only in the catalytic site and the secondary channel in these structures, several models of transcription elongation, based on biochemical studies, propose that template-dependent binding of NTPs in the main channel regulates RNA synthesis. These models, however, remain controversial. We used transient state kinetics and a mutant of RNAP to investigate the role of the main channel in regulating nucleotide incorporation. Our data indicate that a NTP specific for the i + 2 template position can bind to a noncatalytic site and increase the rate of RNA synthesis and that the NTP bound to this site can be shuttled directly into the catalytic site. We also identify fork loop 2, which lies across from the downstream DNA, as a functional component of this site. Taken together, our data support the existence of a noncatalytic template-specific NTP binding site in the main channel that is involved in the regulation of nucleotide incorporation. NTP binding to this site could promote high-fidelity processive synthesis under a variety of environmental conditions and allow DNA sequence-mediated regulatory signals to be communicated to the active site.
Keywords: elongation kinetics, transcription fidelity, allostery, pyrophosphate release, Brownian ratchet
The central role of RNA polymerase (RNAP) in transcription is to catalyze the processive synthesis of the growing RNA transcript with high-fidelity and at reasonable rates. This process is regulated both intrinsically, by RNA and DNA sequence elements, and extrinsically, by nucleotide availability and proteins that interact with the elongating RNAP (1–10). These interactions modulate the conformations of the RNAP ternary elongation complexes (RNAP, DNA, and RNA), which, in turn, affect the rate and fidelity of nucleotide addition and the response of RNAP to regulatory signals. The rate of nucleotide incorporation and the recognition of pause and termination signals by RNAP can be greatly influenced by the sequence of downstream DNA, as well as by the RNA transcript. Subtle changes in the sequence of the downstream DNA can result in dramatic changes in the rates of nucleotide incorporation and the efficiencies of pausing and termination (3, 10–15). The mechanism(s) by which the downstream DNA regulates these processes remains a mystery; however, it has been suggested that nucleoside triphosphates (NTPs) may interact with the downstream DNA to modulate nucleotide incorporation (10, 16, 17).
Crystal structures of prokaryotic and eukaryotic RNAPs reveal two channels: the main channel, which is filled with the downstream DNA, and the secondary channel, which is a negatively charged, funnel-shaped pore that leads from the surface of the enzyme to the active site. NTPs have been observed bound in the catalytic site and secondary channel, but not in the main channel, and there is no obvious path for NTPs to move from the main channel to the active site (18–23). These observations have led to the proposal that the secondary channel is the major, and perhaps only, pathway for NTPs to enter the active site (24–27). Biochemical studies, however, indicate that there may be NTP binding site(s) located in the main channel of RNAP (10, 16, 17). It has been suggested that templated NTPs can bind in the main channel and allosterically enhance the rate of nucleotide incorporation by facilitating translocation (8, 10, 16, 17, 28). In addition, it has been proposed that NTPs may be able to enter the catalytic site via the main channel (8, 10, 16, 17, 29). Currently, however, there is no clear evidence demonstrating that NTPs can even bind to RNAP in the main channel, much less be shuttled from the main channel to the catalytic site, and these proposals remain highly controversial.
To investigate the role, if any, of the main channel in regulating nucleotide incorporation, we characterized the kinetics of incorporation of multiple nucleotides into a growing RNA transcript by performing order-of-addition experiments and by quenching with either EDTA or HCl. Our data indicate that a NTP specific for the i + 2 downstream DNA base may modestly increase the rate of incorporation of the i + 1 NMP and substantially enhances the sequestration of the i + 2 nucleotide and the rate of its subsequent incorporation. In addition, our data demonstrate that NTPs bound to the enzyme prior to the incorporation of the preceding nucleotide can be shuttled into the catalytic site and incorporated into the nascent transcript without being released from the enzyme. Finally, kinetic experiments on a mutant of RNAP suggest that fork loop 2 may comprise part of this downstream NTP binding site, as suggested previously (10, 29).
Results
Using a template in which RNAP is stalled at template position +24 (Fig. 1A), we measured the rates of CMP and AMP incorporation at template positions +25 (i + 1 templated nucleotide) and +26 (i + 2 templated nucleotide), respectively, under two conditions (Fig. 1A): one in which CTP and ATP were added simultaneously and one in which the stalled elongation complexes (SECs) were preincubated with ATP (i + 2) prior to initiating the reaction with the addition of CTP (i + 1). To determine if RNAP sequesters NTPs and to further elucidate the rate-limiting steps of nucleotide incorporation, we used either HCl (denaturing) or EDTA (nondenaturing) to quench the reactions. HCl denatures RNAP; therefore, quenching with HCl measures the amount of product formation (EP + P) at the time of the quench. In contrast, EDTA removes Mg2+ complexed with free NTPs in solution, which renders the NTPs inactive for catalysis; therefore, quenching with EDTA measures the amount of product formation at the time of quench plus any RNAP∶NTP complexes that are able to proceed to product formation without dissociation of the NTP (ES∗ + EP + P) (16, 30, 31). For a single nucleotide addition, quenching with EDTA has been demonstrated to be equivalent to pulse-chase experiments in which unlabeled RNAP elongation complexes are incubated with a radioactively labeled NTP and then an excess of unlabeled NTP is added (32). Comparison of nucleotide incorporation kinetics between these two quenching agents can be used to reveal if there is an accumulation of the enzyme-substrate complex (i.e., RNAP:NTP) prior to phosphodiester bond formation and/or pyrophosphate release (16, 30, 31, 33). If NTP binding and dissociation are in rapid equilibrium, there will be no difference in rates when quenched with EDTA or HCl; however, if the enzyme-substrate complex accumulates prior to bond formation and/or pyrophosphate release, there will be an apparent increase in product formation in the EDTA quench relative to the HCl quench.
Fig. 1.
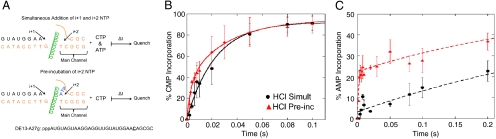
Preincubation of RNAP with the i + 2 NTP increases the rate of bond formation for the i + 2 nucleotide. (A) Schematic of set up for simultaneous and preincubation experiments. The DNA is orange and nascent RNA is black. The bridge helix (green) separates the catalytic site (i + 1) from the main channel site (i + 2). The i + 3 nontemplate nucleotide was omitted for clarity and is not meant to imply that the i + 3 template base is unpaired. (B, C) Plots of the percentage of complexes (normalized to CMP) that have incorporated CMP (i + 1) at template position +25 (B) and AMP (i + 2) at template position +26 (C) for the simultaneous addition (black circles) and preincubation (red triangles) experiments with 100 μM CTP and 10 μM ATP. Reactions were quenched with HCl. Error bars represent the standard deviation in the data. The curves are the best fits to single or double exponentials as determined by statistical F-test.
Preincubation with the i + 2 NTP Increases the Rate of i + 2 Nucleotide Incorporation.
Inspection of the HCl quench data reveals that preincubating the SECs with 10 μM ATP prior to the addition of CTP may slightly increase the rate of CMP incorporation at +25 at early time points (Fig. 1B, Fig. S1A); however, it dramatically increases the rate of AMP incorporation at +26 (Fig. 1C, Fig. S1A). For the simultaneous addition of CTP and ATP, the incorporation of AMP is slow and fits well to a single exponential (2.4 s-1); whereas, preincubation of the SECs with ATP results in the rate of AMP incorporation becoming biphasic (Fig. 1C, Fig. S1A), with the rate of the slow phase being similar to the rate of AMP incorporation in the absence of preincubation (3.0 s-1) (Table S1). These results indicate that preincubating the complexes with the i + 2 NTP increases the fraction of complexes that can rapidly incorporate the i + 2 nucleotide and suggest that the i + 2 NTP (ATP) is binding to RNAP before the incorporation of the i + 1 nucleotide (CMP) to facilitate its own incorporation. To ensure that the observed rate enhancement is not unique to ATP, we performed the same experiments with SECs that were stalled at template position +26, such that the i + 1 and i + 2 positions code for GMP (+27) and CMP (+28), respectively (SI Text, Fig. 1A). Similar to the ATP experiments with RNAP stalled at position +24, we observe a rapid burst of CMP (i + 2) incorporation when the complexes are preincubated with CTP before the addition of GTP relative to when CTP and GTP are added simultaneously (Fig. S2A). We also examined the effect of nonspecific NTPs. Notably, including 1 mM dTTP along with 10 μM ATP during preincubation of SECs at position +24 does not significantly alter CMP (i + 1) incorporation (Fig. S2B) or the rapid burst of AMP (i + 2) incorporation (Fig. S1B, Fig. 2C). Taken together, these data suggest that the enhanced rate of i + 2 incorporation upon preincubation with the i + 2 NTP is specific for the i + 2 NTP and not subject to inhibition by a nonspecific NTP competitor.
Fig. 2.
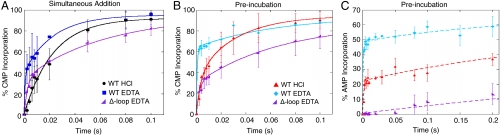
Comparison of nucleotide incorporation for wtRNAP quenched with HCl or EDTA and Δ-loop RNAP quenched with EDTA. Plots of the percentages of complexes (normalized) that have incorporated CMP (i + 1) at template position +25 and AMP at template position +26 for wt and Δ-loop RNAP are shown. (A) CMP incorporation for the simultaneous addition experiments (100 μM CTP and 10 μM ATP) by wtRNAP quenched with HCl (black circles) or EDTA (blue squares) and Δ-loop RNAP quenched with EDTA (purple wedges). (B, C) CMP (B) and AMP (C) incorporation for the preincubation experiments (10 μM ATP prior to the addition of 100 μM CTP) by wtRNAP quenched with HCl (cyan diamonds) or EDTA (red triangles) and Δ-loop RNAP quenched with EDTA (purple wedges). Note that the scale for the x-axis in C is 0.2 s compared to 0.1 s for A and B. Error bars represent the standard deviation in the data. The curves are the best fits to single or double exponentials as determined by statistical F-test.
Preincubation with ATP Leads to Its Rapid Sequestration in the Catalytic Site After CMP Incorporation.
To further examine the mechanism of this rate increase, we also characterized the kinetics of incorporation using an EDTA quench. For CMP incorporation, the fraction of complexes in the burst phase is significantly higher when the complexes are quenched with EDTA relative to being quenched with HCl in both the preincubation and simultaneous addition experiments (Fig. 2 A and B, Fig. S1 A and C). These results indicate that there is an accumulation of enzyme-substrate complex; that is, RNAP sequesters CTP prior to its incorporation. In addition, the rate of the burst phase in the EDTA quench is > 5 times faster than the rate of incorporation in the HCl quench (Table S1), which indicates that the rate-limiting step to incorporation is after sequestration of CTP into the catalytic site. Similar results have been seen with RNAPII and T7 DNA polymerase (17, 28, 34, 35).
For AMP incorporation, there is only a small difference in the rates of incorporation between the HCl and EDTA quenches for the simultaneous addition experiments (Table S1, Fig. S1 A and C, and Fig. S3), indicating that only a small amount of ATP is being sequestered prior to its incorporation when CTP and ATP are added simultaneously. In contrast, for the preincubation experiments, the burst height for AMP incorporation is twofold greater for the EDTA quench relative to the HCl quench, but the rates of the burst and slow phases do not change significantly with the choice of quencher (Table S1, Fig. 2C). The increased burst height in the EDTA quench relative to the HCl quench indicates that preincubating the complexes with ATP prior to initiating synthesis with CTP results in the sequestration of ATP on the enzyme prior to AMP incorporation. In addition, the small difference in the rates of the burst phases under the two quenching conditions (Table S1) suggests that the rate of bond formation is similar to, or faster than, the rate of sequestration and that the difference in burst heights results from rapid bond formation and reversal followed by slow pyrophosphate release (33). Consistent with this suggestion, the HCl and EDTA quench data for the preincubation experiments can be fit by kinetic simulations using a single fast rate constant of 800 s-1 for the formation of the tight (i.e., sequestered) RNAP∶ATP complex, followed by rapid phosphodiester bond formation (600 s-1) and reversal (700 s-1), and a slow rate (3 s-1) of pyrophosphate release (Fig. S4 A and B). In this analysis, the difference in the burst heights between the EDTA and HCl quenches results from rapid reversible bond formation followed by slow pyrophosphate release. Notably, these results and conclusions are similar to those for DNA polymerase I Klenow fragment (33), and a similar equilibrium constant for phosphodiester bond formation (Keq = 600 s-1/700 s-1 = 0.86) was found for T7 DNA polymerase (35). Given that ATP sequestration is partially rate-limiting in the burst phase of AMP incorporation when RNAP is preincubated with ATP, it is likely that the incorporation of AMP is partially rate-limited by pyrophosphate release from the previous incorporation of CMP (see Discussion).
Finally, to ensure that the observed rate enhancements are specific under EDTA quenching, we preincubated the SECs with a nontemplated nucleotide (10 μM GTP) prior to initiating the reaction with CTP and ATP (Fig. S2D). As predicted, we observe no significant increase in the burst of AMP (i + 2) incorporation; however, preincubating with 10 μM GTP (i + 3 nucleotide) slightly increases the rate of the slow phase. Finally, preincubating with 10 μM ATP and 1 mM GTP does not reduce the preincubation effect when the reactions are quenched with EDTA (Fig. S2E). Instead, there is a modest increase in the burst height of the apparent incorporation of AMP. Because GMP (+27) is the next nucleotide to be incorporated after AMP (+26), this increase in burst height may be a result of GTP enhancing the rate of pyrophosphate release after AMP incorporation (Fig. S2E), similar to the results by Johnson et al. (36).
A Deletion Mutant of Fork Loop 2 Ablates Rapid Nucleotide Incorporation.
Based on structural analysis, we previously proposed that fork loop 2, which lies across from the downstream template DNA (Fig. S5) and makes up part of the binding site for the antibiotic streptolydigin (37, 38), may comprise part of a templated NTP binding site (10, 29). Consequently, we generated a mutant RNAP (Δ-loop RNAP) in which four amino acids (βR542–F545) of this loop are deleted (Fig. S5) and characterized the kinetics of nucleotide incorporation using an EDTA quench. Unlike wild type (wt) RNAP, preincubation of Δ-loop RNAP with ATP does not cause an increase in the rate of CMP (i + 1) or AMP (i + 2) incorporation. Instead, deletion of these residues results in a decrease of the apparent rates of CMP and AMP incorporation for both the simultaneous addition (Fig. 2A, Fig. S3) and preincubation experiments (Fig. 2 B and C). These results bolster the proposal that fork loop 2 comprises part of a noncatalytic binding site that binds NTPs in a template-dependent manner. It should be noted, however, that our data cannot distinguish between direct binding of NTPs to fork loop 2 vs. NTPs binding elsewhere on the enzyme and fork loop 2 acting as a functional component.
Further inspection of the data for the preincubation experiments reveals that wtRNAP exhibits biphasic kinetics of AMP incorporation (Fig. 2C). Interestingly, for Δ-loop RNAP, the fast phase is eliminated for AMP incorporation in both the preincubation and simultaneous addition experiments, but the rate of the slow phase (∼1 s-1 for both simultaneous and preincubation) is unaffected (Fig. 2C, Fig. S3, Table S1), suggesting that the slower rate of incorporation is independent of fork loop 2. These results support the existence of at least two pathways for nucleotide incorporation: one that involves fork loop 2 and one that does not. These findings are also consistent with our previous suggestion that fork loop 2 may be part of an allosteric NTP binding site and that nucleotide incorporation follows a nonessential activation mechanism (10, 29, 39).
NTPs Bound in the Noncatalytic Site Can Be Shuttled Directly into the Catalytic Site.
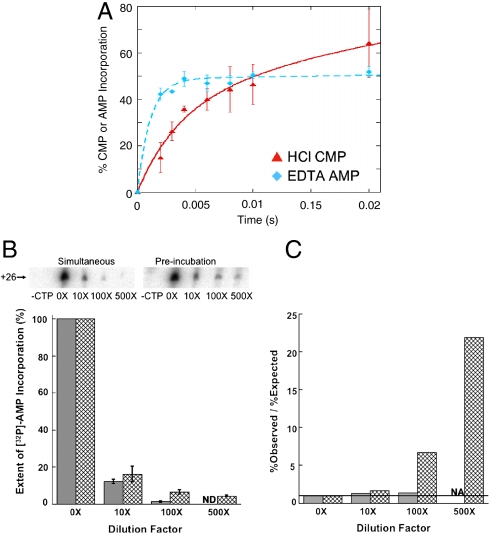
The increased rate of i + 2 nucleotide (AMP) incorporation upon preincubation may result from the i + 2 NTP acting allosterically to open the catalytic site, with a second NTP binding to the catalytic site via the secondary channel and/or from the rapid shuttling of the i + 2 NTP from the noncatalytic site into the catalytic site (8, 10, 16, 17, 29). Comparison of the HCl quench data for CMP (i + 1) incorporation to the EDTA quench data for AMP (i + 2) incorporation can shed light on these possibilities. Notably, the apparent incorporation of AMP in the EDTA experiments (ATP sequestration and AMP bond formation) shows a faster rate than CMP incorporation in the HCl experiments (i.e., CMP bond formation). Inspection of Fig. 3A shows that at 2 ms, only ∼12% of the complexes have incorporated CMP with the HCl quench; whereas, the apparent extent of AMP incorporation is ∼40% with the EDTA quench (Fig. 3A, Fig. S2F). Because the HCl quench measures the extent of nucleotide incorporation, while the EDTA quench measures extent of incorporation and the extent to which sequestered NTPs can be incorporated before dissociating from the enzyme (30, 31, 33), these results indicate that AMP incorporation occurs after the addition of EDTA and demonstrate that ATP∶Mg2+ is sequestered on RNAP and committed to bond formation prior to incorporation of CMP. Taken together, the EDTA and HCl quench data show that RNAP can simultaneously sequester CTP∶Mg2+ and ATP∶Mg2+ prior to incorporation of CMP. Consequently, ATP must be sequestered in a noncatalytic site and then shuttled into the catalytic site, without being released from the enzyme, after CMP incorporation and pyrophosphate release. Importantly, while these results show that ATP can be shuttled from the noncatalytic site to the catalytic site, they do not yield the rate at which ATP is shuttled because incorporation of the sequestered ATP occurs after the EDTA quench.
Fig. 3.
Direct shuttling of the ATP bound in the main channel into the catalytic site upon preincubation of ATP. (A) Comparison of CMP incorporation (100 μM) quenched with HCl (red triangles) to the percent of complexes that have apparently incorporated AMP (10 μM) when quenched with EDTA (cyan diamonds). Data are replotted from Figs. 1B and 2C. (B) Extents of [32P]-AMP incorporation. The representative gels show the amount of labeled transcript at position +26 as a function of the dilution factor ([32P]-ATP/([α-32P]-ATP + ATP)). The bar graph shows the amount of [32P]-AMP incorporation normalized to the zero-fold dilution sample. Solid bars represent the simultaneous addition of CTP and ATP and crosshatched bars represent preincubation of complexes with 10 μM [α-32P]-ATP prior to initiating the reaction with CTP and unlabeled ATP. (C) The bar graph shows the ratio of the observed amount of radioactivity at position +26 relative to the expected amount of radioactivity at +26 based on the dilution factor. The line shows the expected value of the ratio (i) if there is no shuttling of [α-32P]-ATP from noncatalytic site to the catalytic site. The identity of each bar is the same as in (B). Error bars represent the standard deviation in the data. ND means “Not Detected”. NA means “Not Applicable.”
Our data (Fig. 3A, Fig. S2F) support models of nucleotide addition that suggest that NTPs can bind into a site in main channel and subsequently be loaded into the catalytic site (10, 16, 29, 40). To further test this idea, we performed experiments in which unlabeled SECs were preincubated with 10 μM [α-32P]-ATP prior to initiating the reaction with unlabeled ATP and CTP, and then measured the extent of [32P]-AMP incorporation into the transcript as a function of the concentration of unlabeled ATP (Fig. S6, right branch). To account for the incorporation of [32P]-AMP into the transcript that results from [α-32P]-ATP in bulk solution, the same experiment was performed with the simultaneous addition of CTP, 10 μM [α-32P]-ATP, and the corresponding concentration of unlabeled ATP (Fig. S6, left branch). The simultaneous addition of labeled and unlabeled ATP results in the predicted reduction of [32P]-AMP incorporation as a function of the dilution factor ([32P]-ATP/([32P]-ATP + unlabeled ATP)). In contrast, complexes preincubated with 10 μM [α-32P]-ATP prior to the addition of unlabeled CTP and ATP incorporate a significantly higher amount of [32P]-AMP than would be predicted by dilution alone (Fig. 3 B and C), further supporting the conclusion that ATP can bind to a noncatalytic binding site and then be shuttled into the catalytic site and incorporated without being released from the enzyme. The extent of labeling, however, is less than the amount of ATP that appears to be sequestered in a noncatalytic site prior to CMP incorporation (∼30%) and then shuttled into the catalytic site, based on the EDTA experiments (Fig. 3A, Fig. S2F). Furthermore, because the EDTA and HCl experiments indicate that ATP is sequestered in the noncatalytic site (i.e., ATP dissociates slowly from the noncatalytic site), it is unlikely that the lower extent of labeling than expected could be due to exchange of unlabeled ATP with the labeled ATP in the noncatalytic site. Taken together, these observations suggest that shuttling is not the only pathway for NTPs to enter the catalytic site (i.e., NTPs can enter the catalytic site without first binding to the noncatalytic site) and that binding of a templated NTP to the noncatalytic site can allosterically facilitate nucleotide incorporation, with a second NTP subsequently binding into the catalytic site. These conclusions are consistent with our previous observation showing that incubation of SECs with a high concentration of the nonincorporatable ATP analog AMPCPP significantly increases the rate of incorporation of AMP at the i + 2 position (39).
Discussion
Several models have been proposed for nucleotide binding and incorporation by RNAP. These models fit into two classes: simple Brownian ratchet models, in which RNAP is in an equilibrium between the pre- and posttranslocated states and a single NTP binds to the catalytic site in the posttranslocated state via the secondary channel (5, 41–43), and NTP-facilitated incorporation models, in which there is a second NTP binding site where templated NTPs bind and facilitate catalytic site opening, pyrophosphate release, and/or translocation (8, 10, 16, 17, 28, 39). Two recent reviews present evidence that indicates that a simple Brownian ratchet is insufficient to explain the mechanism of nucleotide incorporation catalyzed by RNAP. These reviews put forth similar NTP-facilitated incorporation models (8, 29). Both models include the binding of a templated NTP in a noncatalytic binding site in the main channel, and both suggest that the NTP bound in this site may be shuttled directly into the catalytic site; however, until now, there was no clear evidence that two NTPs could simultaneously bind RNAP or that a NTP could bind in a noncatalytic site and be shuttled to the catalytic site.
Binding of the i + 2 NTP Likely Facilitates Pyrophosphate Release from Incorporation of the i + 1 NMP.
As discussed above, comparison of the EDTA and HCl quench data for AMP incorporation when ATP (i + 2) is preincubated indicates that a step at, or before, NTP sequestration is partially rate-limiting for the fast phase. This observation suggests that pyrophosphate release from the previous incorporation of CMP (i + 1) may be rate-limiting and that binding of ATP to the noncatalytic site may facilitate pyrophosphate release. Given that pyrophosphate must be released after the incorporation of CMP (i + 1) before ATP can bind to the catalytic site, our results suggest that i + 2 NTP can bind to an noncatatytic binding site on RNAP prior to the incorporation of the i + 1 nucleotide and facilitate the release of pyrophosphate and opening of the catalytic site, thus allowing the i + 2 nucleotide to rapidly enter the catalytic site. This suggestion is supported by studies on Escherichia coli RNAP that demonstrated that the presence of 1 mM of the i + 2 NTP (but not nonspecific NTPs) increases the rate of pyrophosphate release from the i + 1 incorporation event by 200-fold (from a rate of ∼3 s-1 in the absence of the i + 2 NTP to ∼600 s-1 in the presence of the i + 2 NTP) (36). Notably, the rates of AMP incorporation in our HCl quench experiments (Table S1) are similar to the unassisted and NTP-assisted rates of pyrophosphate release observed by Johnson et al., suggesting that pyrophosphate release may be a rate-limiting step to nucleotide incorporation during processive synthesis (16, 36). Notably, enhancement of pyrophosphate release by the i + 2 NTP could also explain the modest increase in the rate of CMP (i + 1) incorporation when ATP is preincubated before the addition of CTP (Fig. 1B, Table S1).
The Pretranslocated State of RNAP May Represent the Fast State.
It has been generally accepted that the posttranslocated state with the catalytic site accessible via the secondary channel was responsible for the rapid incorporation by RNAP; however, recent observations that two fidelity mutants of yeast RNAP II (rpb1-E1103A and rpb9-Δ) exhibit increased rates of nucleotide incorporation (both correct and incorrect), even though they are shifted to the pretranslocated state relative to wtRNAP, have brought this idea into question (8, 29, 34, 44). After incorporation of a terminating nucleotide, these RNAP mutants shift heavily towards the pretranslocated state, but incubation of the complexes with the next templated nucleotide shifts the complexes towards the posttranslocated state. This observation has been interpreted as RNAP being in an equilibrium between the pre- and posttranslocated states and binding of the templated NTP to the catalytic site in the posttranslocated state acting as a pawl (5, 34, 44), which it likely does. This suggestion, however, does not account for the observation that these mutant RNAPs are shifted to the pretranslocated state relative to wtRNAP but incorporate nucleotides more rapidly than wtRNAP. Our observation that occupancy of the i + 2 NTP in the noncatalytic site when RNAP is in the pretranslocated state increases i + 2 NMP incorporation suggest a possible alternative explanation for this effect. The next incoming NTP may bind to the template-dependent noncatalytic site when RNAP is in the pretranslocated state and facilitate the transition into the posttranslocated state (10). Our data, together with the data on these fidelity mutants, suggest that rapid nucleotide incorporation may result from binding of the next templated NTP to the noncatalytic site when RNAP is in the pretranslocated state followed either by a second NTP binding into the catalytic site or shuttling of the NTP from the noncatalytic to the catalytic site, and that the posttranslocated (or pretranslocated) state with the noncatalytic site unoccupied may be responsible for the slow phase.
Models of Transcription Elongation.
We recently proposed a facilitated Brownian ratchet model for nucleotide incorporation in which there are three possible pathways to nucleotide incorporation: a nonallosteric path, an allosteric path, and a shuttle path (29). In the nonallosteric pathway, the noncatalytic site is not utilized, and NTPs enter the catalytic site directly when RNAP is in the posttranslocated state, probably via the secondary channel. We suggest that this path corresponds to the slow phase of the biphasic kinetics of incorporation, as well as incorporation by the Δ-loop RNAP mutant. Two plausible structural explanations for slow incorporation on the nonallosteric path are that in the absence of a NTP bound in the noncatalytic site, (i) pyrophosphate release and/or trigger loop opening may be slow, blocking entry of the next NTP into the catalytic site, or (ii) closing of the trigger loop may be slow resulting in slow incorporation. Such scenarios may come into play when complexes are stalled or paused.
In both the shuttle and allosteric pathways, we have proposed that a templated NTP binds to a site in the main channel and facilitates pyrophosphate release and/or opening of the catalytic site after the previous incorporation event and may facilitate sequestration of the NTP into the catalytic (29). As discussed above, our EDTA and HCl quench data are consistent with NTP binding in the noncatalytic site facilitating pyrophosphate release and NTP sequestration, as well as shuttling of NTPs. In addition, our current and previous data (39) suggest that a second NTP can bind into the catalytic site after one binds in the noncatalytic site. Notably our data do not unequivocally locate the noncatalytic site in the main channel; however, our data, as well as those of previous studies showing effects of the i + 2 NTP on nucleotide incorporation, strongly support the suggestion that the noncatalytic site is in the main channel (10, 16, 28, 45). First, in all of the published crystal structures of RNAP elongation complexes, the i + 2 template base is located in the main channel (18–23, 43, 46). Second, our observation that the noncatalytic site can be occupied by the i + 2 NTP during incorporation of the i + 1 NTP makes it highly unlikely that the noncatalytic site could be located in the secondary channel, because the trigger loop, which is believed to accompany catalysis (47, 48), blocks the secondary channel upon closing. Third, we identify fork loop 2, which lies across from the downstream DNA, as a functional component of this allosteric site.
For the shuttle pathway, there appear to be two possible routes by which NTPs could move from the main channel to the catalytic site (29): over the top of the bridge helix along with the template, as proposed by Burton and coworkers (16), or under the bridge helix via the trigger loop as it closes. It has been suggested that the allosteric pathway could be explained without a noncatalytic site by template misalignment such that the i + 2 template base transiently occupies the catalytic site with the i + 1 nucleotide flipped out (49, 50). This suggestion was based on the observation that the i + 2 nucleotide was preferentially misincorporated in RNAP complexes containing only a primer and template. If such a mechanism were occurring in our studies, we would expect to see significant misincorporation of AMP for CMP in the preincubation experiments; however, we do not see any misincorporation of AMP (i + 2) for CMP (i + 1) in these studies or in our studies of misincorporation, even at concentrations as high as 1 mM ATP (9). In addition, Burton and coworkers found that high concentrations of the i + 2 NTP reduces misincorporation at i + 1 for RNAP II, which is in direct contradiction to the predictions of the misalignment model (16). Notably, even in the studies on which the misalignment model is based, very little misincorporation of the i + 2 nucleotide was observed when the RNAP complexes were assembled with a nontemplate strand as well as the primer and template (49). Taken together, these results argue strongly against template misalignment as an explanation for the allosteric effect. It has also been suggested that binding of NTPs into the “E site,” which is in the secondary channel and overlaps with the catalytic site, could shift the equilibrium between pre- and posttranslocated states to the posttranslocated state and thereby enhance incorporation (51). Given that NTPs in the E site do not interact with the template base, binding of NTPs to the E site cannot explain our data or any of the previous data showing template-dependent effects of NTPs on the enhancement of nucleotide incorporation. Finally, neither of these models could account for having two templated NTPs bound to RNAP simultaneously.
The overall role that this noncatalytic binding site plays in elongation remains unknown. The affinity of NTPs for the noncatalytic site or whether the site is utilized likely depends on sequence-dependent conformations of SECs. The noncatalytic site potentially could be involved in pause site recognition and escape, as well as in transcription fidelity. Consistent with a role in pausing, pausing efficiencies are increased at low NTP concentrations (14, 52), where ternary complexes might be more susceptible to regulatory signals due to low occupancy of the templated NTP in the noncatalytic site (7). Interestingly, the efficiency of pausing can be highly dependent on the identity of the nucleotide at the i + 2 position after the site of RNAP pausing (15). In addition, the rate of escape from a his pause site exhibits a nonhyperbolic NTP concentration-dependence (48), consistent with two NTPs being involved in pause escape, and it was suggested that movement of βD-loopII, which forms the “top” of the catalytic site, leads to movements in fork loop 2 via an allosteric mechanism during pausing (48). The presence of a secondary NTP binding site also may permit RNAP to maintain high-fidelity processive synthesis under a variety of environmental conditions while still being able to respond to regulatory signals. For example, shuttling of NTPs may provide a double check on nucleotide identity: checking once in the main channel and again in the catalytic site (16, 17), which could significantly increase transcriptional fidelity under nonoptimal conditions, such as an imbalance of NTP pools. Taken together, our data suggest that the noncatalytic site may serve as a template-dependent sensor of NTPs to regulate transcription elongation.
Materials and Methods
Enzyme and Templates.
wt his6-tagged RNAP was purified from E. coli strain RL916 (gift of R. Landick) using the protocol described previously (53, 54). The DE13-A27g template DNA was prepared as previously described (10).
Transcription Elongation Experiments.
The formation and purification of SECs is described in the SI Text and elsewhere (10, 39). Presteady-state kinetic experiments were conducted on a RQF-3 rapid quench device (Kintek Corporation). All NTP concentrations are reported as final working concentrations.
Data Quantification and Normalization.
The quantification of the radioactivity at each position was performed as previously described (10, 39). To compare different datasets, the data were normalized based on the highest percentage of complexes that incorporated to position +25 (CMP) and longer, as described in detail previously (39). Briefly, the percentage of complexes that reached positions +25 (CMP) and +26 (AMP) were divided by the highest percentage of complexes that reached +25, thereby normalizing CMP incorporation to 100% and AMP incorporation to the extent of CMP incorporation. Data for the EDTA experiments are the average of three or four experiments. The HCl experiments are averages of two trails for each condition. The data for the NTP loading experiments were quantified by dividing the amount of radioactivity at transcript position +26 in each gel lane by the amount of radioactivity in the 0-fold dilution reaction. The reported data are the average of three trials. For more detailed information on the methods used, see SI Text.
Supplementary Material
Acknowledgments.
We thank L. Spremulli, T. Kunkel, M. Kireeva, Z. Burton, and T. Santangelo for useful comments and suggestions. We thank T. Santangelo and R. Landick for providing reagents. This work was funded by National Institutes of Health (NIH) grants GM 79480 and GM 54316.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011274108/-/DCSupplemental.
References
- 1.von Hippel P, Yager T. Transcript elongation and termination are competitive kinetic processes. Proc Nat Acad Sci USA. 1991;88:2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Hippel P, Yager T. The elongation-termination decision in transcription. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Nat Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 5.Bar-Nahum G, et al. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 6.Boukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Nat Acad Sci USA. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erie DA. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim Biophys Acta. 2002;1577:224–239. doi: 10.1016/s0167-4781(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 8.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. BBA- Gene Regul Mech. 2010;1799:389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel P. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 10.Holmes SF, Erie DA. Downstream DNA sequence effects on transcription elongation: Allosteric binding of nucleoside triphosphates facilitates translocation via a ratchet motion. J Biol Chem. 2003;278:35597–35608. doi: 10.1074/jbc.M304496200. [DOI] [PubMed] [Google Scholar]
- 11.Lee DN, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- 12.Reynolds R, Bermbdez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase: I. Analysis of 13 Rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 13.Herbert KM, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palangat M, Hittinger CT, Landick R. Downstream DNA selectively affects a paused conformation of human RNA polymerase II. J Mol Biol. 2004;341:429–442. doi: 10.1016/j.jmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Chan CL, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site: mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–20804. [PubMed] [Google Scholar]
- 16.Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Burton ZF. A tunable ratchet driving human RNA polymerse II translocation adjusted by accurately templated nucleoside triphosphates loaded at downstream sites and by elongation factors. J Biol Chem. 2007;282:36582–36592. doi: 10.1074/jbc.M707014200. [DOI] [PubMed] [Google Scholar]
- 18.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 19.Kettenberger H, Armache K-J, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 20.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 21.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Nat Acad Sci USA. 2004;101:17361–17364. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide Microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 27.Adelman K, et al. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol Cell. 2004;14:753–762. doi: 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Nedialkov YA, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- 29.Erie DA, Kennedy SR. Forks, pincers, and triggers: the tools for nucleotide incorporation and translocation in multi-subunit RNA polymerases. Curr Opin Struct Biol. 2009;19:708–714. doi: 10.1016/j.sbi.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochem. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold JJ, Gohara DW, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochem. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kireeva M, et al. Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods. 2009;48:333–345. doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochem. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 34.Kireeva ML, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel SS, Wong I, Johnson KA. Presteady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochem. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 37.Ho MX, Hudson BP, Das K, Arnold E, Ebright RH. Structures of RNA polymerase-antibiotic complexes. Curr Opin Struct Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuske S, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 40.Burton ZF, et al. NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem Cell Biol. 2005;83:486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- 41.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat Struct Mol Biol. 2008;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, et al. Structural basis of transcription: backtracked RNA polymerase II at 3.4 Angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walmacq C, et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA Polymerase II. J Biol Chem. 2009;284:19601–19612. doi: 10.1074/jbc.M109.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Zobeck KL, Burton ZF. Human RNA polymerase II elongation in slow motion: role of the TFIIF RAP74 a1 helix in nucleoside triphosphate-driven translocation. Mol Cell Biol. 2005;25:3583–3595. doi: 10.1128/MCB.25.9.3583-3595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sydow JF, et al. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Kaplan CD, Larsson K-M, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Kashkina E, et al. Template misalignment in multisubunit RNA polymerases and transcription fidelity. Mol Cell. 2006;24:257–266. doi: 10.1016/j.molcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Pomerantz RT, Temiakov D, Anikin M, Vassylyev DG, McAllister WT. A mechanism of nucleotide misincorporation during transcription due to template-strand misalignment. Mol Cell. 2006;24:245–255. doi: 10.1016/j.molcel.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Temiakov D, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 52.Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J Mol Biol. 2001;311:265–282. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- 53.Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochem. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 54.Uptain SM, Chamberlin MJ. Escherichia coli RNA polymerase terminates transcription efficiently at Rho-independent terminators on single-stranded DNA templates. Proc Nat Acad Sci USA. 1997;94:13548–13553. doi: 10.1073/pnas.94.25.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.