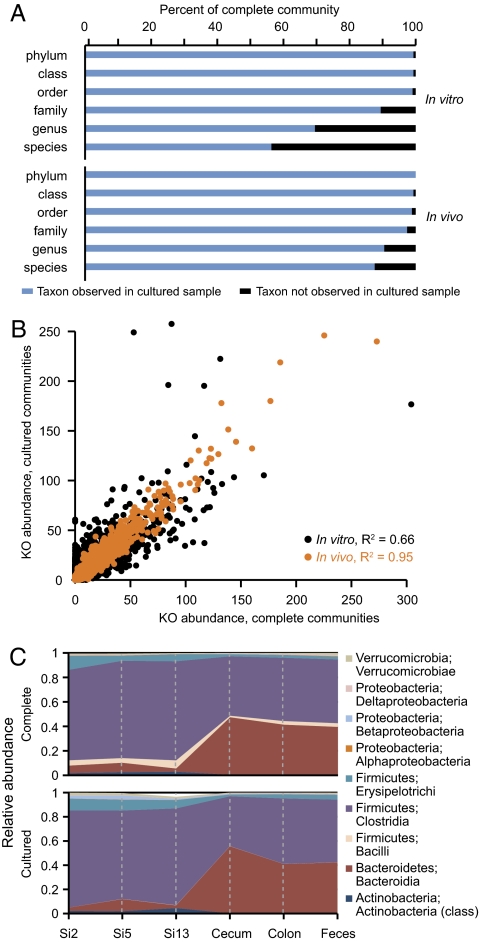
Fig. 1.
Comparison of the taxonomic representation of bacterial species and gene content in complete versus cultured human fecal microbial communities before and after their introduction into gnotobiotic mice. (A) 16S rRNA sequences from complete microbiota were compared with those identified from microbial communities cultured from the same human donors. At each taxonomic level, the proportion of reads in the complete community belonging to a taxonomic group observed in the cultured sample is shown in blue; the proportion of reads belonging to a taxonomic group not observed in the cultured sample (or lacking taxonomic assignment) is shown in black. Data shown are the average of two unrelated human donors. In vitro samples refer to comparisons between human fecal samples and plated material. In vivo samples refer to comparisons between gnotobiotic mice colonized with a complete human fecal microbiota and mice colonized with the readily cultured microbes from the same human fecal sample. (B) Annotated functions identified in the microbiomes of complete and cultured human gut communities. Each point represents a KO designation plotted by relative abundance (average across two donors, per 100,000 sequencing reads). Black points represent KO comparisons between the in vitro samples; orange points represent comparisons between in vivo samples. (C) The distribution of taxa and their relative abundance along the length of the intestine are similar in gnotobiotic mice colonized with complete or cultured human gut communities. Relative abundances of class-level taxa at six locations are shown; data represent the average of mice colonized from two unrelated donors. Si, small intestine divided into 16 equal-size segments and sampled at Si-2 (proximal), Si-5 (middle), and Si-13 (distal). PCoA suggests that gut biogeography, rather than donor or culturing, explains the majority (58%) of variance between samples (Fig. S3 A–C).