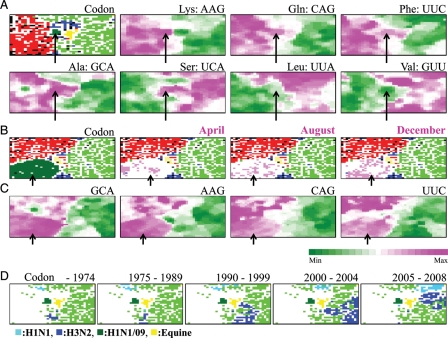
Figure 3.
Codon-BLSOM. (A) Codon: BLSOM was constructed for synonymous codon usage in approximately 34 000 genes from influenza A strains including H1N1/09 isolated in the early stage (April and May), as described by Kanaya et al.,21 and lattice points were indicated in a colour representing the host as described in Fig. 1B. Occurrence levels of seven codons, which were diagnostic for host-specific separation, were indicated with different levels of two colours: pink (high), green (low), and achromatic (intermediate) as described by Kanaya et al.21 Because the synonymous codon usage was analysed, two codons belonging to each two-codon box gave the complementary patterns, and therefore, the pattern of one codon was listed for those belonging to two-codon boxes. (B) Codon: BLSOM was constructed for synonymous codon usage in approximately 60 000 genes from influenza A strains including all H1N1/09strains, and lattice points were indicated in a colour representing the host as described in Fig. 1B. (C) Occurrence levels of four diagnostic codons in the Codon-BLSOM presented in (B) were indicated as described in (A). Scale for the occurrence levels in (A) and (C) was indicated in the colours shown at the bottom of (C). (D) Retrospective time-series changes for human subtype strains (H1N1 and H3N2) on the Codon-BLSOM presented in (A). A zone for H1N1/09 or equine strains was additionally marked for reference to help recognize the position in Codon-BLSOM in Fig. 1A. Each strain was indicated in the colours shown at the bottom of (D).