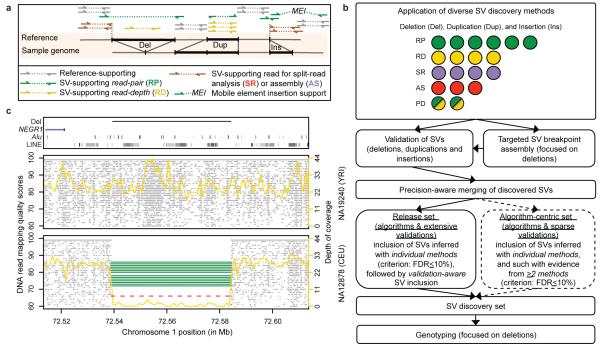
Figure 1. SV discovery and genotyping in population scale sequence data.
A. Schematic depicting the different modes (i.e., approaches) of sequence based SV detection we used. The RP approach assesses the orientation and spacing of the mapped reads of paired-end sequences14,15 (reads are denoted by arrows); the RD approach evaluates the read depth-of-coverage25,26; the SR approach maps the boundaries (breakpoints) of SVs by sequence alignment28,29; the AS approach assembles SVs30,31,32. B. Integrated pipeline for SV discovery, validation, and genotyping. Colored circles represent individual SV discovery methods (listed in Supplementary Table 1), with modes indicated by a color scheme: green=RP; yellow=RD; purple=SR; red=AS; green and yellow=methods evaluating RP and RD (abbreviated as ‘PD’). C. Example of a deletion, previously associated with BMI35, identified independently with RP (green), RD (yellow), and SR (red) methods. Grey dots indicate position and mapping quality for individual sequence reads. Targeted assembly confirmed the breakpoints detected by SR.