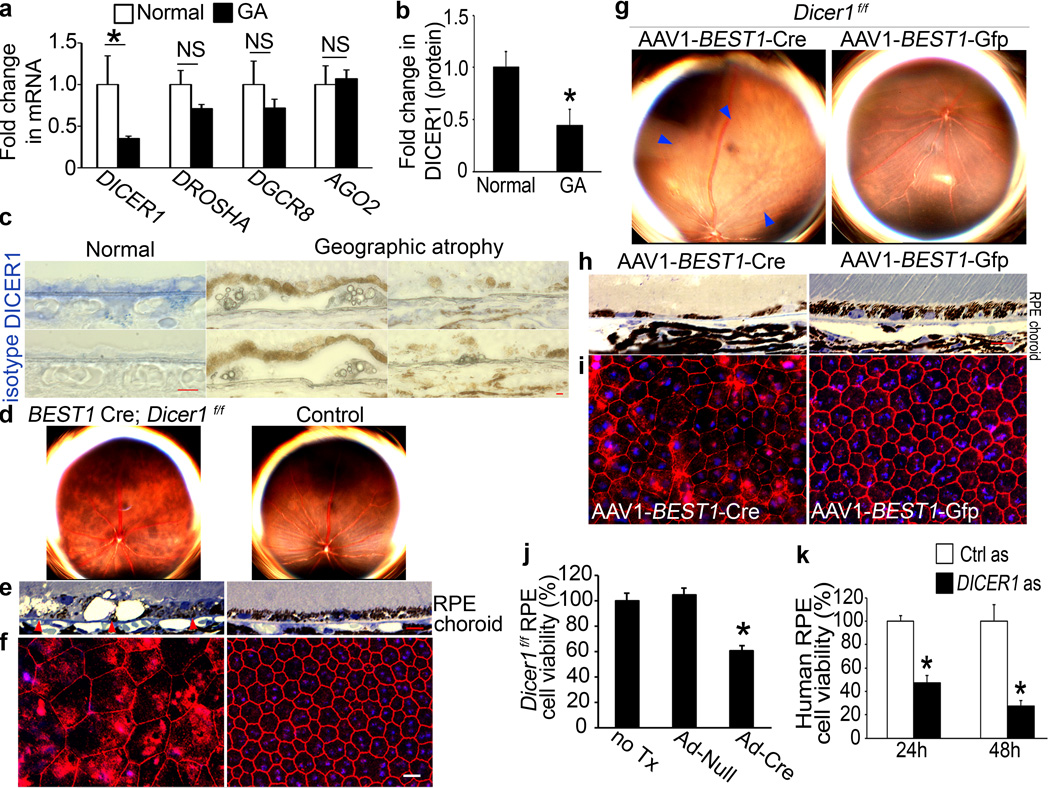
Figure 1. DICER1 deficit in GA induces RPE degeneration.
a, DICER1 less abundant in RPE of human eyes with GA (n=10) compared to control RPE (n=11). P = 0.004 by Mann Whitney U test. DROSHA, DGCR8, and EIF2C2 (encoding AGO2) abundance not significantly different (P > 0.11 by Mann Whitney U test). n=10–11. b, DICER1 quantification, assessed by Western blotting (Supplementary Fig. 1), lower in human GA RPE (n=4) compared to control RPE (n=4). P = 0.003 by Student t test. c, Reduced DICER1 (blue) in human GA RPE compared to control eyes. d, e, Fundus photographs (d) and toluidine-blue-stained sections (e) show RPE degeneration in BEST1 Cre; Dicer1f/f mice but not controls. Arrowheads point to basal surface of RPE. f, Flatmounts stained for zonula occludens-1 (ZO-1; red) show RPE disruption in BEST1 Cre; Dicerf/f mice compared to controls. g, h, Fundus photographs (g) and toluidine blue-stained sections (h) show RPE (g, h) and photoreceptor (h) degeneration in Dicer1f/f mice following subretinal injection of AAV1-BEST1-Cre but not AAV1-BEST1-GFP. i, Flatmounts show Dicer1f/f mouse RPE degeneration following subretinal injection of AAV1-BEST1-Cre but not AAV1-BEST1-GFP. Nuclei stained blue with Hoechst 33342. Representative images shown. n=16–32 (d–f); 10–12 (g–i). Scale bars, (c,e,h), 10 µm; (f,i) 20 µm. j, Adenoviral vector coding for Cre recombinase (Ad-Cre) treatment reduces Dicer1f/f mouse RPE cell viability compared to Ad-Null or untreated (no Tx) cells. k, DICER1 antisense (as) reduces human RPE cell viability compared to control antisense (Ctrl as)-treated cells. n=6–8. All error bars indicate mean±s.e.m.