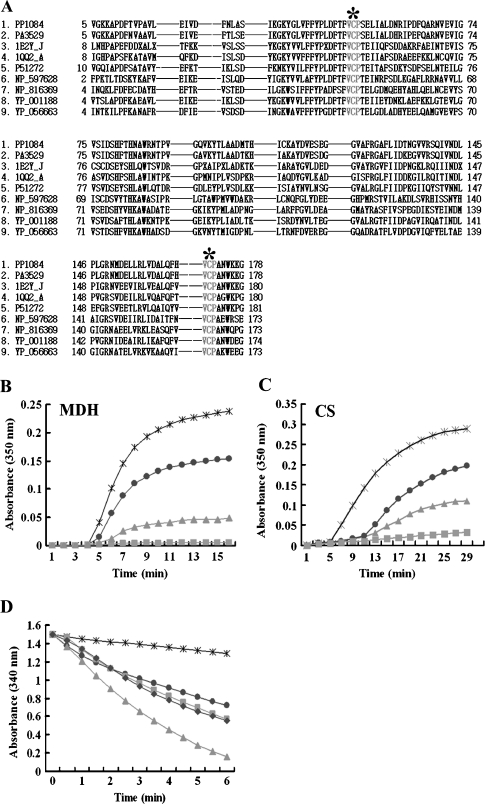
Fig. 2.
Investigation of PpPrx enzymatic functions. a Alignment of the amino acid sequences of P. putida PpPrx (2-Cys Prx) with homologous AhpC and Prxs from several representative prokaryotes and eukaryotes. The encoded amino acid sequences were aligned, and gaps (dashes) were introduced to optimize the sequence alignment. Two highly conserved VCP tripeptides, which are related to the catalytic function, are designated by gray letters and asterisks. The abbreviations for the amino acid sequences of Prxs from various species are as follows: 1 PP1084 of P. putida KT2440, 2 PA3529 of P. aeruginosa PAO1, 3 1E2Y_J of Crithidia fasciculata, 4 1QQ2_A of Rattus norvegicus, 5 P51272 of Porphyra purpurea, 6 NP_597628 of Encephalitozoon cuniculi GB-M1, 7 NP_816369 of Enterococcus faecalis, 8 YP_001188 of Leptospira sp., and 9 YP_056663 of Propionibacterium acnes. b The chaperone activity of recombinant PpPrx was measured using aggregation of MDH at 43°C at different molar ratios: 1 MDH to 0 PpPrx (asterisk), 1 MDH to 0.1 PpPrx (dots), 1 MDH to 0.5 PpPrx (squares), and 1 MDH to 1 yeast TPx (triangles). c The chaperone activity of recombinant PpPrx was measured using aggregation of CS at 43°C at different molar ratios: 1 CS to 0 PpPrx (asterisk); 1 CS to 0.2 PpPrx (dots); 1 CS to 0.5 PpPrx (triangles), and 1 CS to 1 PpPrx (squares). d Peroxide reductase activity of recombinant PpPrx from P. putida was measured with the yeast Trx system at different concentrations: without PpPrx (asterisk) in the reaction buffer; 10 μM PpPrx (dots); 20 μM PpPrx (squares); 40 μM PpPrx (triangle); and 5 μM yeast TPx (diamond; control). The data shown are the means of at least three independent experiments