Abstract
We investigated associations of early pregnancy maternal peripheral blood gene expression with preeclampsia. In a nested case control study, gene expression of peripheral blood, collected at 16weeks of gestation on average from 16 women destined to develop preeclampsia and 16 women who had normotensive pregnancies was profiled using Affymetrix GeneChip Arrays. Fold change and Student's T-test analyses were used to compare differential gene expression across the groups. Functions and functional relationships as well as common regulatory sequences of differentially expressed genes were investigated. Genes participating in abnormal placentation (e.g COL1A1), immune/inflammation response (e.g. IKBKB) and cellular development (including cell cycle) (e.g. RBI) were differentially expressed in early pregnancy peripheral blood in preeclampsia. We identified transcription factors (i.e. Sp1, MAZ and MZF1) that may account for co-expression of differentially expressed genes. Preeclampsia is associated with differential gene expression in early pregnancy peripheral blood.
Keywords: preeclampsia, early pregnancy, gene, expression
Introduction
The pathogenesis of preeclampsia, a pregnancy-related vascular disorder, is a complex process that has been associated with angiogenesis, immune dysfunction, inflammation and oxida-tive stress [1-3]. While preeclampsia is a disorder of the second half of pregnancy, accumulating evidence supports the multi-stage developmental phases of preeclampsia that start early in pregnancy [1-3]. For instance, immune sensitivity and abnormal placentation in early pregnancy contribute to placental hypoxemia which promotes diffuse inflammation, oxidative stress and endothelial dysfunction later in pregnancy [1-4]. However, significant gaps in knowledge persist on preeclampsia related events and risk factors in early pregnancy that is critical for prevention and early detection of disease [5].
Increasingly, gene expression studies are being used to investigate pathophysiologic processes underlying preeclampsia [5]. Several investigators, including our team, have conducted gene expression profiling of preeclamptic placenta after delivery [6-9]. Although results from these studies provide new insights about preeclampsia pathophysiology, inferences are limited by critical questions concerning temporal relationships between gene expression profiles, onset of the clinical disorder, and its management. Few gene expression studies investigating preeclampsia were conducted in early pregnancy [10-12] and even fewer were conducted using early pregnancy peripheral blood [12], a tissue that may reflect local and systemic pathophysi-ological changes associated with preeclampsia.
Taking into account the potential significance of this research area, in 2003, we expanded an on -going pregnancy cohort study by prospectively collecting and storing peripheral blood samples in Paxgene™ Blood RNA tubes for gene expression studies. In this report, we describe findings of a nested case control study that investigated early pregnancy maternal peripheral blood gene expressions among 16 women destined to develop preeclampsia and 16 women who had normotensive pregnancies. We also compared similarities and differences between preeclampsia related underlying pathomechanisms in early and late pregnancy using gene expression profiles of peripheral blood (early pregnancy) and placenta (at-delivery), respectively.
Materials and methods
Study population
This nested case control study was conducted using information collected from participants of the Omega study (1996-2007), a prospective study designed to examine risk factors of pregnancy complications. Participants were recruited from women who initiate prenatal care before 20 weeks gestation at Swedish Medical Center (SMC) affiliated clinics. Ineligibility criteria included < 18 years of age, not speaking or reading English, not planning to carry the pregnancy to term, and/or not planning to deliver at SMC. The study for this report was conducted among selected preeclampsia cases (N=16) and controls (N=16) from Omega cohort members enrolled during the period of July 2003 to May 2007. During this interval, > 80% of approached women consented to participate in the study and > 95% of enrolled participants were followed through pregnancy completion.
Preeclampsia cases were selected using the then current 1996 ACOG guidelines when both pregnancy-induced hypertension (PIH) and proteinuria were present. PIH was defined as a sustained (≥2 measures 6 hrs apart) blood pressure (Bp) elevation (>140/90 mmHg) after 20 weeks of gestation or a sustained 15-mm Hg rise in diastolic Bp or a 30-mm Hg rise in systolic Bp above 1st trimester values. Proteinuria was defined as a sustained (≥2 measures 4 hrs apart) presence of elevated protein in the urine (>30 mg/dL or >1+on a urine dipstick). Controls were selected among women who had normotensive pregnancies uncomplicated by proteinuria or gestational diabetes. Women who were multiparous or had history of chronic hypertension and/or pre-gestational diabetes as well as women with non-singleton pregnancies were excluded. The Institutional Review Board of the SMC approved study protocols. All participants provided written informed consent.
Data collection
Information on risk factors was collected using in-person interviews, blood collection and medical records abstraction. Following enrollment, in -person interviews were conducted to collect data on socio-demographic characteristics and reproductive and medical histories. At or near the time of in-person interviews (16 weeks of gestation on average), trained phlebotomists collected peripheral blood samples. PAXgene™ Blood RNA tubes (Qiagen Inc, Valencia, CA) [13] were used to collect blood samples for gene expression studies. After delivery, trained personnel abstracted maternal and infant medical records to ascertain pregnancy outcomes.
Total RNA extraction, target preparation and hybridization
The PAXgene Blood RNA Kit (Qiagen Inc., Valencia, CA) was used for extraction and purification of total RNA. Total RNA concentration was determined by UV absorbance at 260 nm (A260) by direct measurement on a NanoDrop ND1000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE). RNA purity was assessed by evaluating readings at 260 nm and 280 nm (A260/ A280). All samples had A260/ A280 values > 2.0 indicating high level of purity. Samples were then kept in frozen storage at -80°C. All RNA samples, including reference RNAs, underwent quality control checks and were labeled using same standardized protocols.
RNA target preparations were conducted using guidelines of the NuGEN™ Ovation™ RNA Amplification System V2 (amplification) and the NuGEN™ FL-Ovation™ cDNA Biotin Module V2 (fragmentation and labeling) (NuGen Technologies Inc., San Carlos, CA). The resultant fragmented and labeled cDNA was added to the hybridization cocktail in accordance with the NuGEN and Affymetrix guidelines for hybridization onto Affymetrix Human Genome U133 Plus 2.0 GeneChip® Arrays (Affymetrix, Sunnyvale, CA). The arrays were washed and stained on the GeneChip® Fluidics Station 450 (Affymetrix, Sunnyvale, CA), before being inserted into the Affymetrix autoloader carousel and scanned using the GeneChip® Scanner 3000 (Affymetrix, Sunnyvale, CA). Data from each array was quantified using GeneChip® Operating Software (Affymetrix, Sunnyvale, CA).
GeneChip quality controls and normalization
GeneChip quality control procedures included the following. First, background values of GeneArray scanners calibrated to the new PMT setting (10% of maximum) were assessed for comparability. Second, GAPDH gene was used to assess RNA sample and assay quality. Third, controls on the GeneChip array (four E.Coli genes, bioB, bioC and bioD and the cre gene) were spiked into each sample to evaluate hybridization efficiency. Fourth, raw noise (Q value), a measure of pixel-to-pixel variation of probe cells due to operation-associated electrical noise, was evaluated. Fifth, PolyA control genes (dap, lys, phe, thr and trp genes from B. subtilis) were amplified and spiked into the RNA samples prior to amplification to serve as internal control genes. Finally, data were normalized using an error-weighted model based on Rosetta Resolver Error Models (Rosetta, Seattle, WA)[14].
Real time quantitative polymerase chain reaction (RT-qPCR) experiment
RT-qPCR experiment was conducted to confirm microarray based expression measures of selected genes. Initially, 1 μg total RNA was reverse transcribed using the Transcriptor first strand cDNA synthesis kit (Roche Applied Science, Indianapolis, IN). The qPCR reactions were performed using the Roche LightCycler 480® Probes kit and the LightCycler 480® instrument (Roche Applied Science, Indianapolis, IN). Pre-designed exon spanning Taqman® assays for each gene target were obtained from Applied Biosystems (Foster City, CA). Each individual assay was run on an individual 96-well plate in duplicate for each sample; and, 2 reverse transcription negative controls and 2 no template control wells were included with each assay. Individual reactions were characterized by the PCR cycle at which fluorescence first rises above threshold background fluorescence (the threshold cycle, Ct). ACTB and GAPDH genes, selected based on their non-variant gene expression across cases and controls in the microarray experiment, were used for normalization.
Statistical analysis
Analysis was conducted on normalized and log-transformed data. Fold change (FC) expression differences (absolute FC ≥ 1.5) and Student's T-test (two sample, unequal variances) p-values (<0.05) were used to identify differentially expressed genes across the two groups (cases and controls). Two-Dimensional hierarchical clustering, using Cluster and TreeView softwares [16], and Principle components analysis (PCA) techniques were used to evaluate whether differentially expressed genes cluster arrays into groups (case and control groups) [15]. Functions and functional relationships between differentially expressed genes were investigated using Ingenuity Pathway Analysis (IPA), as described before (Ingenuity, Redwood City, CA) [7]. Gene-enrichment of networks (network score) based on a modified Fisher's exact test, measured in IPA, was used to rank biological significance of gene function networks in relation to preeclampsia. In the confirmatory RT-qPCR experiment, we used fold change analysis and Student's T-test to compare whether results were consistent with those obtained from microarray experiments. Common regulatory sequences for the differentially expressed genes as well as their cognate regulators (transcription factors (TFs)) were searched using ConTra (conserved transcription factor binding sites, TFBs) and MAPPER [17]. Finally, using GeneGO pathway analysis tools (GeneGO Inc., St Joseph, MI), we compared gene ontology (GO) processes represented by differentially expressed genes in maternal early pregnancy peripheral blood in the current study with differentially expressed genes in preeclamptic placenta we reported before [7].
Results
Selected study population characteristics are summarized in Table 1. Mean age of preeclampsia cases and normotensive controls were 35.1 and 32.1 years, respectively. Maternal whole blood samples were collected from participants at 16 weeks of gestation, on average. Preeclampsia cases had higher pre-gestational BMI compared with controls.
Table 1.
Characteristics of study population
| Characteristics | Preeclampsia cases (N = 16) | Normotensive controls (N=16) |
|---|---|---|
| GA at blood collection, weeks* | 16.2 (1.7) | 16.2 (2.5) |
| Maternal Age, years* | 35.1 (5.3) | 32.1(4.4) |
| 20-34 years | 8 (50.0) | 13 (81.3) |
| 35 and above years | 8 (50.0) | 3(13.7) |
| Maternal Race/Ethnicity | ||
| White | 14(87.5) | 13 (81.3) |
| African American | 2 (12.5) | 1 (6.3) |
| Other | 0 (0.0) | 2 (12.5) |
| Pre-gestational BMI, kg/m2* | 29.6 (11.9) | 23.8 (6.2) |
| <20 | 2 (12.5) | 1 (6.3) |
| 20-24.99 | 7 (43.8) | 12 (75.0) |
| 25-29.99 | 5 (31.3) | 2 (12.5) |
| ≥30 | 2 (12.5) | 1 (6.3) |
| Smoked in pregnancy | 0 (0.0) | 1 (6.3) |
| Family history of chronic hypertension | 10 (62.5) | 6(37.5) |
| Family history of diabetes mellitus | 3 (18.8) | 1 (6.3) |
| Gestational diabetes | 2 (12.5) | 0 (0.0) |
Mean (standard deviation), otherwise n (%). Abbreviations: GA: gestational age, BMI: body mass index; kg/m2: kilogram/meter2
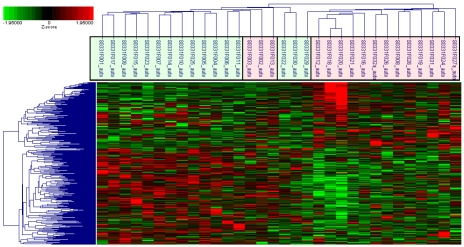
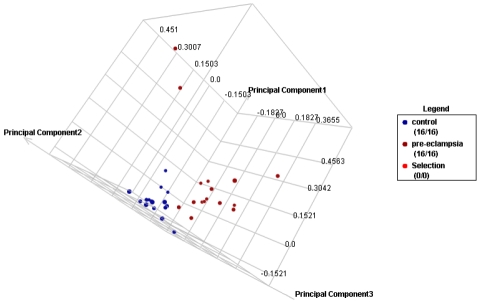
Of the total >38,500 genes represented by ∼47,400 probe sets on the GeneChip, 247 genes (<1%) represented by 356 probes that met the following criteria were up (N=86) or down (N=161) regulated in preeclampsia cases compared to controls; Student's T-test p-value < 0.05 and absolute fold change > 1.5 (Table 2 and 3). These differentially expressed genes included genes involved in abnormal placentation (e.g. COL1A1 and NRTK2) and immune response/inflammation (e.g. CLEC12B and IKBKB). The range of fold change differences in expression between preeclampsia cases and controls was -5.40 (DKFZp666G057) to 2.78 (TMEM176B). In hierarchical clustering, based on expressions measured by probes representing differentially expressed genes, all but three preeclampsia cases and all but one normotensive controls clustered in to the two main cluster groups of cases and controls, respectively (Figure 1). Similarly, in PCA, we demonstrated that preeclampsia cases and controls can be classified into two groups using expressions measured by probes representing differentially expressed genes (Figure 2).
Table 2.
Selected* list of differentially expressed genes
| Gene Symbol | Gene Name | FC* | P-value* |
|---|---|---|---|
| Down regulated genes | |||
| DKFZp666G057 | hypothetical protein DKFZp666G057 | -5.40 | 0.0245 |
| HSD17B12 | Hydroxysteroid (17-beta) dehydrogenase 12 | -3.39 | 0.0121 |
| PLEKHG2 | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | -2.79 | 0.0157 |
| COL5A3 | collagen, type V, alpha 3 | -2.69 | 0.0432 |
| LOC400581 | GRB2-related adaptor protein-like | -2.64 | 0.0037 |
| ACCN2 | amiloride-sensitive cation channel 2, neuronal | -2.38 | 0.0078 |
| GTSF1L | gametocyte specific factor 1-like | -2.35 | 0.0033 |
| CLEC12B | C-type lectin domain family 12, member B | -2.33 | 0.0010 |
| COL1A1 | collagen, type I, alpha 1 | -2.26 | 0.0466 |
| ZNF496 | zinc finger protein 496 | -2.21 | 0.0003 |
| VN1R1 | vomeronasal 1 receptor 1 | -2.13 | 0.0006 |
| PTPRM | protein tyrosine phosphatase, receptor type, M | -2.05 | 0.0012 |
| IKBKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | -2.02 | 0.0124 |
| PTPRM | protein tyrosine phosphatase, receptor type, M | -2.02 0.0003 | |
| Up regulated genes | |||
| NTRK2 | neurotrophic tyrosine kinase, receptor, type | 2 2.01 | 0.0185 |
| LOC728806 | Similar to N-ethylmaleimide-sensitive factor | 2.10 | 0.0081 |
| MSL-1 | Male-specific lethal-1 homolog | 2.10 | 0.0018 |
| VEPH1 | ventricular zone expressed PH domain homolog 1 (zebrafish) | 2.24 | 0.0107 |
| B9D1 | B9 protein domain | 1 2.41 | 0.0106 |
| MGC50559 | hypothetical protein MGC50559 | 2.41 | 0.0493 |
| RAB6A | RAB6A, member RAS oncogene family | 2.50 | 0.0159 |
| NALCN | sodium leak channel, non-selective | 2.67 | 0.0022 |
| PTPRD | protein tyrosine phosphatase, receptor type, D | 2.67 | 0.0106 |
| TMEM176B | Transmembrane protein 176B | 2.78 | 0.0046 |
Selected (absolute fold change > 2.0 [FC] and Student's T test p-value [p-value] < 0.05) list of differentially expressed genes.
Table 3.
List of differentially expressed genes
| Gene Symbol | Gene Name | FC* | P-value* |
|---|---|---|---|
| DKFZp666G057 | hypothetical protein DKFZp666G057 | -5.4 | 0.0245 |
| HSD17B12 | Hydroxysteroid (17-beta) dehydrogenase 12 | -3.39 | 0.0121 |
| PLEKHG2 | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | -2.79 | 0.0157 |
| COL5A3 | collagen, type V, alpha 3 | -2.69 | 0.0432 |
| LOC400581 | GRB2-related adaptor protein-like | -2.64 | 0.0037 |
| ACCN2 | amiloride-sensitive cation channel 2, neuronal | -2.38 | 0.0078 |
| GTSF1L | gametocyte specific factor 1-like | -2.35 | 0.0033 |
| CLEC12B | C-type lectin domain family 12, member B | -2.33 | 0.0010 |
| COL1A1 | collagen, type I, alpha 1 | -2.26 | 0.0466 |
| ZNF496 | zinc finger protein 496 | -2.21 | 0.0003 |
| VN1R1 | vomeronasal 1 receptor 1 | -2.13 | 0.0006 |
| PTPRM | protein tyrosine phosphatase, receptor type, M | -2.05 | 0.0012 |
| IKBKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | -2.02 | 0.0124 |
| PLXNA1 | plexin A1 | -1.99 | 0.0040 |
| LTK | leukocyte tyrosine kinase | -1.96 | 0.0268 |
| ULK4 | unc-51-like kinase 4 (C. elegans) | -1.96 | 0.0160 |
| AGRN | agrin | -1.94 | 0.0293 |
| GIMAP5 | GTPase, IMAP family member 5 | -1.93 | 0.0027 |
| RAB40A | RAB40A, member RAS oncogene family | -1.93 | 0.0187 |
| CLEC12A | C-type lectin domain family 12, member A | -1.92 | 0.0023 |
| DKFZP761N09121 | hypothetical protein DKFZp761N09121 | -1.9 | 0.0050 |
| RAB3IP | RAB3A interacting protein (rabin3) | -1.89 | 0.0002 |
| MUC5B | mucin 5B, oligomeric mucus/gel-forming | -1.88 | 0.0019 |
| PICK1 | protein interacting with PRKCA 1 | -1.88 | 0.0282 |
| FAM70A | family with sequence similarity 70, member A | -1.87 | 0.0275 |
| DUSP2 | dual specificity phosphatase 2 | -1.86 | 0.0000 |
| LOC161527 | promyelocytic leukemia | -1.86 | 0.0209 |
| PTGDS | prostaglandin D2 synthase 21kDa (brain) | -1.85 | 0.0004 |
| MLZE | melanoma-derived leucine zipper, extra-nuclear factor | -1.83 | 0.0018 |
| UACA | uveal autoantigen with coiled-coil domains and ankyrin repeats | -1.83 | 0.0481 |
| MTHFSD | Methenyltetrahydrofolate synthetase domain containing | -1.82 | 0.0484 |
| TTC28 | tetratricopeptide repeat domain 28 | -1.82 | 0.0011 |
| LRRC23 | leucine rich repeat containing 23 | -1.81 | 0.0094 |
| CA3 | carbonic anhydrase III, muscle specific | -1.8 | 0.0155 |
| CENTG2 | centaurin, gamma 2 | -1.8 | 0.0003 |
| GPR4 | G protein-coupled receptor 4 | -1.8 | 0.0011 |
| LDB2 | LIM domain binding 2 | -1.8 | 0.0204 |
| SOX15 | SRY (sex determining region Y)-box 15 | -1.8 | 0.0436 |
| GIPC3 | GIPC PDZ domain containing family, member 3 | -1.79 | 0.0189 |
| LYPD3 | LY6/PLAUR domain containing 3 | -1.79 | 0.0081 |
| DSTN | Destrin (actin depolymerizing factor) | -1.78 | 0.0015 |
| KLHDC4 | Kelch domain containing 4 | -1.78 | 0.0176 |
| PCOLCE | procollagen C-endopeptidase enhancer | -1.77 | 0.0132 |
| ZNF542 | zinc finger protein 542 | -1.77 | 0.0418 |
| FLJ44606 | hypothetical gene supported by AK126569 | -1.75 | 0.0093 |
| FAM120AOS | family with sequence similarity 120A opposite strand | -1.74 | 0.0191 |
| HEL308 | DNA helicase HEL308 | -1.74 | 0.0057 |
| HLCS | holocarboxylase synthetase (biotin-(proprionyl-Coenzyme Acarboxylase (ATP-hydrolysing)) ligase) | -1.74 | 0.0044 |
| TMEM46 | transmembrane protein 46 | -1.74 | 0.0064 |
| TRIM47 | tripartite motif-containing 47 | -1.74 | 0.0119 |
| CASP10 | caspase 10, apoptosis-related cysteine peptidase | -1.73 | 0.0124 |
| CEL | carboxyl ester lipase (bile salt-stimulated lipase) | -1.73 | 0.0300 |
| GPRASP2 | G protein-coupled receptor associated sorting protein 2 | -1.73 | 0.0018 |
| RDH16 | retinol dehydrogenase 16 (all-trans) | -1.73 | 0.0262 |
| DHCR7 | 7-dehydrocholesterol reductase | -1.72 | 0.0033 |
| FN1 | fibronectin 1 | -1.72 | 0.0145 |
| PQLC3 | PQ loop repeat containing 3 | -1.72 | 0.0219 |
| USP18 | ubiquitin specific peptidase 18 | -1.71 | 0.0485 |
| LOC150837 | hypothetical protein LOC150837 | -1.7 | 0.0311 |
| TMEM177 | transmembrane protein 177 | -1.7 | 0.0084 |
| LOC283859 | hypothetical protein LOC283859 | -1.69 | 0.0039 |
| TAPBP | TAP binding protein (tapasin) | -1.69 | 0.0481 |
| LOC283666 | hypothetical protein LOC283666 | -1.68 | 0.0059 |
| HEYL | hairy/enhancer-of-split related with YRPW motif-like | -1.67 | 0.0283 |
| SERHL | serine hydrolase-like | -1.67 | 0.0073 |
| ZCCHC2 | zinc finger, CCHC domain containing 2 | -1.67 | 0.0118 |
| ACOT4 | acyl-CoA thioesterase 4 | -1.66 | 0.0020 |
| C10orf58 | chromosome 10 open reading frame 58 | -1.66 | 0.0079 |
| CXCR6 | chemokine (C-X-C motif) receptor 6 | -1.66 | 0.0017 |
| MKL2 | MKL/myocardin-like 2 | -1.66 | 0.0356 |
| OSGIN1 | oxidative stress induced growth inhibitor 1 | -1.66 | 0.0496 |
| ZNF804A | zinc finger protein 804A | -1.66 | 0.0091 |
| ARL3 | ADP-ribosylation factor-like 3 | -1.65 | 0.0011 |
| GPA33 | glycoprotein A33 (transmembrane) | -1.65 | 0.0215 |
| LOC751071 | hypothetical protein LOC751071 | -1.65 | 0.0043 |
| RNF157 | CDNA FLJ36181 fis, clone TESTI2026794 | -1.65 | 0.0176 |
| SF3A2 | splicing factor 3a, subunit 2, 66kDa | -1.65 | 0.0075 |
| 7A5 | putative binding protein 7a5 | -1.64 | 0.0016 |
| PTPN20A | protein tyrosine phosphatase, non-receptor type 20B | -1.64 | 0.0396 |
| HEMK1 | HemK methyltransferase family member 1 | -1.63 | 0.0053 |
| PMS2L4 | postmeiotic segregation increased 2-like 4 | -1.63 | 0.0252 |
| TDRKH | tudor and KH domain containing | -1.63 | 0.0028 |
| CD248 | CD248 molecule, endosialin | -1.62 | 0.0186 |
| FLJ35934 | FLJ35934 protein | -1.62 | 0.0322 |
| OIP5 | Opa interacting protein 5 | -1.62 | 0.0125 |
| C5orf20 | chromosome 5 open reading frame 20 | -1.61 | 0.0312 |
| FLJ45224 | FLJ45224 protein | -1.61 | 0.0083 |
| HDAC5 | histone deacetylase 5 | -1.61 | 0.0031 |
| PDZD4 | PDZ domain containing 4 | -1.61 | 0.0231 |
| ATG9B | ATG9 autophagy related 9 homolog B (S. cerevisiae) | -1.6 | 0.0183 |
| CASKIN2 | CASK interacting protein 2 | -1.6 | 0.0006 |
| FBXO15 | F-box protein 15 | -1.6 | 0.0250 |
| FLJ37512 | similar to Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3) | -1.6 | 0.0307 |
| GRAMD1B | GRAM domain containing 1B | -1.6 | 0.0112 |
| LOC25845 | hypothetical LOC25845 | -1.6 | 0.0208 |
| LOC791120 | zinc finger protein 783 | -1.6 | 0.0180 |
| LRRC56 | leucine rich repeat containing 56 | -1.6 | 0.0102 |
| ZNF10 | zinc finger protein 10 | -1.6 | 0.0210 |
| C3orf39 | chromosome 3 open reading frame 39 | -1.59 | 0.0016 |
| CYP4V2 | Cytochrome P450, family 4, subfamily V, polypeptide 2 | -1.59 | 0.0027 |
| HEY2 | hairy/enhancer-of-split related with YRPW motif 2 | -1.59 | 0.0168 |
| PMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) | -1.59 | 0.0192 |
| TMEM132E | transmembrane protein 132E | -1.59 | 0.0224 |
| TRIM46 | tripartite motif-containing 46 | -1.59 | 0.0301 |
| CACNB1 | calcium channel, voltage-dependent, beta 1 subunit | -1.58 | 0.0096 |
| OBSCN | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | -1.58 | 0.0085 |
| ANKS6 | ankyrin repeat and sterile alpha motif domain containing 6 | -1.57 | 0.0279 |
| BNIPL | BCL2/adenovirus E1B 19kD interacting protein like | -1.57 | 0.0137 |
| JAG2 | jagged 2 | -1.57 | 0.0007 |
| KPTN | kaptin (actin binding protein) | -1.57 | 0.0245 |
| ME3 | malic enzyme 3, NADP(+)-dependent, mitochondrial | -1.57 | 0.0172 |
| MPI | mannose phosphate isomerase | -1.57 | 0.0098 |
| RNF12 | Ring finger protein 12 | -1.57 | 0.0122 |
| CLEC11A | C-type lectin domain family 11, member A | -1.56 | 0.0059 |
| COL23A1 | collagen, type XXIII, alpha 1 | -1.56 | 0.0188 |
| DOCK4 | dedicator of cytokinesis 4 | -1.56 | 0.0169 |
| GARNL3 | GTPase activating Rap/RanGAP domain-like 3 | -1.56 | 0.0018 |
| SPHK2 | sphingosine kinase 2 | -1.56 | 0.0170 |
| TMEM117 | transmembrane protein 117 | -1.56 | 0.0128 |
| ZNF668 | zinc finger protein 668 | -1.56 | 0.0325 |
| CARD11 | caspase recruitment domain family, member 11 | -1.55 | 0.0076 |
| CHD9 | chromodomain helicase DNA binding protein 9 | -1.55 | 0.0428 |
| COL6A1 | collagen, type VI, alpha 1 | -1.55 | 0.0209 |
| H2AFY | H2A histone family, member Y | -1.55 | 0.0459 |
| IGSF11 | immunoglobulin superfamily, member 11 | -1.55 | 0.0236 |
| TCF19 | transcription factor 19 (SC1) | -1.55 | 0.005 |
| UBQLNL | ubiquilin-like | -1.55 | 0.0101 |
| C5orf42 | chromosome 5 open reading frame 42 | -1.54 | 0.0055 |
| IFRG15 | interferon responsive gene 15 | -1.54 | 0.0142 |
| LPHN1 | latrophilin 1 | -1.54 | 0.0052 |
| MGC15705 | hypothetical protein MGC15705 | -1.54 | 0.0451 |
| NRCAM | neuronal cell adhesion molecule | -1.54 | 0.0055 |
| PTCH1 | patched homolog 1 (Drosophila) | -1.54 | 0.019 |
| RASGRP3 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | -1.54 | 0.0029 |
| RUNX2 | runt-related transcription factor 2 | -1.54 | 0.0089 |
| USP43 | ubiquitin specific peptidase 43 | -1.54 | 0.0057 |
| CSNK1E | casein kinase 1, epsilon | -1.53 | 0.0191 |
| CST4 | cystatin S | -1.53 | 0.0195 |
| DKFZP434C153 DKFZP434C153 protein | -1.53 | 0.0022 | |
| EDIL3 | EGF-like repeats and discoidin I-like domains 3 | -1.53 | 0.0341 |
| FGFR4 | fibroblast growth factor receptor 4 | -1.53 | 0.0189 |
| LOC388963 | similar to short-chain dehydrogenase/reductase 1 | -1.53 | 0.0094 |
| NOS3 | nitric oxide synthase 3 (endothelial cell) | -1.53 | 0.0032 |
| C15orf50 | chromosome 15 open reading frame 50 | -1.52 | 0.0179 |
| C2orf40 | chromosome 2 open reading frame 40 | -1.52 | 0.0257 |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | -1.52 | 0.0131 |
| GATA3 | GATA binding protein 3 | -1.52 | 0.0022 |
| PRR6 | Proline rich 6 | -1.52 | 0.0314 |
| SPON1 | spondin 1, extracellular matrix protein | -1.52 | 0.0025 |
| CACHD1 | cache domain containing 1 | -1.51 | 0.0404 |
| MCOLN3 | mucolipin 3 | -1.51 | 0.0115 |
| PCDH7 | protocadherin 7 | -1.51 | 0.0042 |
| PPP1R13L | protein phosphatase 1, regulatory (inhibitor) subunit 13 like | -1.51 | 0.0134 |
| SLC24A1 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | -1.51 | 0.0332 |
| SNRP70 | small nuclear ribonucleoprotein 70kDa polypeptide (RNP antigen) | -1.51 | 0.0392 |
| ATP13A4 | ATPase type 13A4 | -1.5 | 0.0369 |
| DCST2 | DC-STAMP domain containing 2 | -1.5 | 0.0173 |
| DMPK | dystrophia myotonica-protein kinase | -1.5 | 0.0204 |
| INADL | InaD-like (Drosophila) | -1.5 | 0.0394 |
| SNAPC4 | small nuclear RNA activating complex, polypeptide 4, 190kDa | -1.5 | 0.0075 |
| TMEM182 | transmembrane protein 182 | -1.5 | 0.0346 |
| GOLIM4 | golgi integral membrane protein 4 | 1.5 | 0.0372 |
| GOLM1 | golgi membrane protein 1 | 1.5 | 0.0253 |
| TGM4 | transglutaminase 4 (prostate) | 1.5 | 0.0011 |
| ADAM3A | ADAM metallopeptidase domain 3A (cyritestin 1) | 1.51 | 0.0185 |
| BCAS1 | breast carcinoma amplified sequence 1 | 1.51 | 0.0066 |
| HIST1H2BG | histone cluster 1, H2bg | 1.51 | 0.0206 |
| MGC13005 | hypothetical protein MGC13005 | 1.51 | 0.0056 |
| KBTBD2 | kelch repeat and BTB (POZ) domain containing 2 | 1.52 | 0.0034 |
| MUC20 | Mucin 20, cell surface associated | 1.52 | 0.0142 |
| E2F1 | E2F transcription factor 1 | 1.53 | 0.0431 |
| CENPN | centromere protein N | 1.54 | 0.0259 |
| HUS1B | HUS1 checkpoint homolog b (S. pombe) | 1.54 | 0.0096 |
| MIPOL1 | mirror-image polydactyly 1 | 1.54 | 0.0192 |
| LAPTM4B | lysosomal associated protein transmembrane 4 beta | 1.55 | 0.0211 |
| LOC728142 | hypothetical protein LOC728142 | 1.55 | 0.0059 |
| PAP2D | phosphatidic acid phosphatase type 2 | 1.56 | 0.0227 |
| SAMD5 | sterile alpha motif domain containing 5 | 1.56 | 0.0492 |
| WNK3 | WNK lysine deficient protein kinase 3 | 1.56 | 0.0234 |
| PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 1.57 | 0.0023 |
| TMOD2 | tropomodulin 2 (neuronal) | 1.57 | 0.0392 |
| CDH10 | cadherin 10, type 2 (T2-cadherin) | 1.58 | 0.0316 |
| GAS2L3 | growth arrest-specific 2 like 3 | 1.58 | 0.0409 |
| HIPK1 | homeodomain interacting protein kinase 1 | 1.58 | 0.0336 |
| LOC151877 | hypothetical protein LOC151877 | 1.58 | 0.0136 |
| LONRF2 | LON peptidase N-terminal domain and ring finger 2 | 1.58 | 0.0412 |
| CTD-2248C21.2 | G antigen 1 | 1.59 | 0.0415 |
| AK7 | adenylate kinase 7 | 1.61 | 0.0114 |
| TFRC | transferrin receptor (p90, CD71) | 1.61 | 0.0081 |
| TMC1 | transmembrane channel-like 1 | 1.61 | 0.0216 |
| ABCC4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 1.62 | 0.0052 |
| HIST1H4E | histone cluster 1, H4e | 1.62 | 0.0038 |
| PAK7 | p21(CDKN1A)-activated kinase 7 | 1.62 | 0.0326 |
| EGLN3 | egl nine homolog 3 (C. elegans) | 1.63 | 0.0009 |
| XYLB | xylulokinase homolog (H. influenzae) | 1.63 | 0.0219 |
| CDC20B | Cell division cycle 20 homolog B (S. cerevisiae) | 1.64 | 0.0428 |
| RBI | retinoblastoma 1 (including osteosarcoma) | 1.64 | 0.0001 |
| SPAG4L | sperm associated antigen 4-like | 1.64 | 0.0176 |
| ARHGEF12 | Rho guanine nucleotide exchange factor (GEF) 12 | 1.65 | 0.0201 |
| TSPAN17 | tetraspanin 17 | 1.65 | 0.0146 |
| CHD7 | chromodomain helicase DNA binding protein 7 | 1.66 | 0.0204 |
| IGSF3 | immunoglobulin superfamily, member 3 | 1.66 | 0.0187 |
| KIAA0644 | KIAA0644 gene product | 1.66 | 0.0167 |
| DDX54 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | 1.67 | 0.0069 |
| LTB4DH | leukotriene B4 12-hydroxydehydrogenase | 1.67 | 0.0416 |
| DCBLD2 | discoidin, CUB and LCCL domain containing 2 | 1.68 | 0.01 |
| KRT33A | keratin 33A | 1.68 | 0.0312 |
| PSG4 | pregnancy specific beta-1-glycoprotein 4 | 1.68 | 0.0371 |
| KIAA0746 | KIAA0746 protein | 1.69 | 0.0034 |
| MFAP5 | microfibrillar associated protein 5 | 1.71 | 0.0238 |
| OR2B2 | olfactory receptor, family 2, subfamily B, member 2 | 1.71 | 0.0457 |
| SCN3B | sodium channel, voltage-gated, type III, beta | 1.71 | 0.0493 |
| LOC283194 | hypothetical protein LOC283194 | 1.72 | 0.0499 |
| SHC4 | SHC (Src homology 2 domain containing) family, member 4 | 1.72 | 0.0146 |
| SRGAP2P1 | SLIT-ROBO Rho GTPase activating protein 2 pseudogene 1 | 1.72 | 0.0454 |
| TAT | Tyrosine aminotransferase | 1.72 | 0.0366 |
| KRT25 | keratin 25 | 1.73 | 0.0084 |
| ATP13A3 | ATPase type 13A3 | 1.74 | 0.0463 |
| TTC7A | Tetratricopeptide repeat domain 7A | 1.74 | 0.0178 |
| EMCN | endomucin | 1.75 | 0.0254 |
| TMEM63A | Transmembrane protein 63A | 1.75 | 0.0300 |
| EVI1 | Ecotropic viral integration site 1 | 1.77 | 0.0365 |
| KCNB2 | potassium voltage-gated channel, Shab-related subfamily, member 2 | 1.77 | 0.0333 |
| FLJ14959 | hypothetical protein FLJ14959 | 1.78 | 0.0072 |
| UNC119B | Unc-119 homolog B (C. elegans) | 1.78 | 0.0352 |
| PHTF2 | putative homeodomain transcription factor 2 | 1.79 | 0.0159 |
| UTP11L | UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) | 1.8 | 0.0496 |
| DEFB107A | defensin, beta 107A | 1.81 | 0.0363 |
| LRP6 | low density lipoprotein receptor-related protein 6 | 1.85 | 0.0161 |
| RNF32 | ring finger protein 32 | 1.86 | 0.0377 |
| KIAA2022 | KIAA2022 | 1.87 | 0.0241 |
| ANKRD30B | ankyrin repeat domain 30B | 1.88 | 0.0446 |
| EDN1 | endothelin 1 | 1.91 | 0.0163 |
| RNF150 | ring finger protein 150 | 1.91 | 0.0197 |
| MMP25 | matrix metallopeptidase 25 | 1.95 | 0.0191 |
| PDE1A | phosphodiesterase 1A, calmodulin-dependent | 1.96 | 0.0244 |
| OR5H1 | olfactory receptor, family 5, subfamily H, member 1 | 1.98 | 0.0429 |
| NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 2.01 | 0.0185 | |
| LOC728806 | Similar to N-ethylmaleimide-sensitive factor 2.1 | 0.0081 | |
| MSL-1 | Male-specific lethal-1 homolog 2.1 | 0.0018 | |
| VEPH1 | ventricular zone expressed PH domain homolog 1 (zebrafish) 2.24 | 0.0107 | |
| B9D1 | B9 protein domain 1 2.41 | 0.0106 | |
| MGC50559 | hypothetical protein MGC50559 2.41 | 0.0493 | |
| RAB6A | RAB6A, member RAS oncogene family 2.5 | 0.0159 | |
| NALCN | sodium leak channel, non-selective 2.67 | 0.0022 | |
| PTPRD | protein tyrosine phosphatase, receptor type, D 2.67 | 0.0106 | |
| TMEM176B | Transmembrane protein 176B 2.78 | 0.0046 |
List of differentially expressed genes in fold change (FC) and Students' T-test (p-value) analyses.
Figure 1.
Hierarchical clustering of participants and differentially expressed genes. Probes (N=356) representing differentially expressed genes (N=247) (upregulated: shades of red and downregulated: shades of green) (rows) and participants (columns, cases=pink and controls=green) grouped according to level and nature of expression and similarity of expression profiles (participants) and subjected to hierarchical tree clustering.
Figure 2.
Principal components analysis. Principal component analysis results of all samples (16 preeclampsia cases and 16 controls) using expressions measured by probes (N=356) representing differentially expressed genes (N=247). (Red/ right: cases, Blue/left: controls).
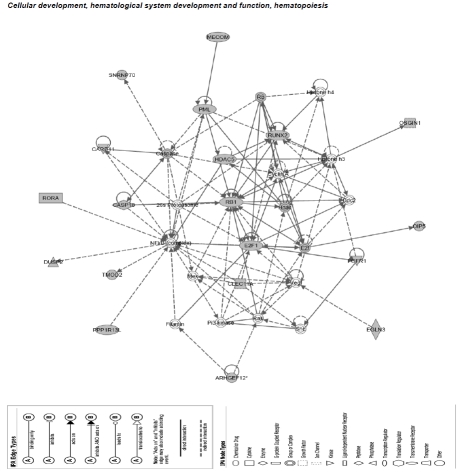
We further evaluated functions and functional relationships of differentially expressed genes. In IPA, 12 networks with network scores > 3 were over represented by differentially expressed genes. These networks are involved in cellular development (particularly of the hematological system), cell signaling, cell cycle regulation, metabolism (lipid, vitamin, carbohydrates and nucleic acids), inflammation and cellular response (Table 4). In particular, the RBI_ E2F1 cell cycle pathway that regulates cellular development (e.g in the hematological system) was significantly over represented (Figure 3).
Table 4.
Gene networks overrepresented by differentially expressed genes
| # | Genes in Network* | Score | Focus genes | Functions |
|---|---|---|---|---|
| 1 | 26s Proteasome, ARHGEF12, CARD11, CASP10, Caspase, Cdc2, CLEC11A, Cyclin A, DUSP2, E2f, E2F1, EGLN3, FGFR1, Filamin, Hdac, HDAC5, Histone h3, Histone h4, MECOM, Mek, NFkB (complex), OIP5, OSGIN1, Pi3-kinase, PML, PPP1R13L, Ras, Rb, RBI, RORA, RUNX2, She, SNRNP70, TM0D2, Vegf | 32 | 19 | Cellular Development, Hematological System Development and Function, Hematopoiesis |
| 2 | CA3, CD28, CENPN, COL23A1, CSGALNACT1, CSNK1E, CSNK1G2, dihydrotestosterone, EPS15, GRB2, HEYL, KCNB2, LRP6, LRRC23, NAA38, ONECUT1, PMS1, PPP1R3D, PPP2R1A, PRNP, RDH5, RDH16, REPS1, RNF20, RPS28, SGIP1, SLC24A1, SMAD3, SOST, SVIL, TGM4, TMF1, UGT2B11, UGT2B15, UGT2B® | 22 | 14 | Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism |
| 3 | AGAP1, Alp, Apl, COL5A3, Collagen type I, Collagen(s), DCBLD2, EDN1, ERK, ERK1/2, Fgf, FGFR4, FN1, Focal adhesion kinase, HEY2, Ifn gamma, I LI, Laminin, LYPD3, Mapk, MUC5B, NTRK2, PCDH7, PCOLCE, Pdgf, PDGF BB, PI3K, Pkc(s), PLC gamma, PLEKHG2, Rac, Rapl, RASGRP3, TCR, Tgf beta | 22 | 14 | Cellular Development, Visual System Development and Function, Cellular Assembly and Organization |
| 4 | Actin, Actin-Nrf2, BCL2, BNIPL, CD248, DSTN, EMCN, ENDOG, FMO1, F0XF2, GFI1B, GIMAP5, GSTT1, JAG2, JARID2, LGALS3BP, MCOLN3, MEGF6, MEGF8, MFAP5, MYH14, NCALD, NFE2L2, NQ02, PDZD4, SERPINB8, SLC1A4, SNAPC4, STARD3, TBP, TNF, TROPONIN, UACA, Vacuolar H+ ATPase, ZNF496 | 20 | 13 | Cell Morphology, Cell-To-Cell Signaling and Interaction, Cell Death |
| 5 | CASKIN2, CDH6, CDH7, CDH8, CDH9, CDH10, CDH15, CDH17, CDH18, CDH22, CLYBL, CTNNAL1, CTNNB1, EDIL3, FBX08, FBX015, GNB2L1, GOLM1, GPX2, Groucho, HLCS, HNF1A, KRT33A, MIRLET7D (includes EG:406886), MKL2, NRCAM, PCCA, PTCH1, Scf Trcp beta, SKP1, SRF, TRIM46, TSG101, TSPAN17, ZNF365 | 19 | 13 | Cell-To-Cell Signaling and Interaction, Tissue Development, Embryonic Development |
| 6 | ARL3, C100RF58, CD70, CEL, CST4, DGKA, GIP2, GPA33, Hla-abc, IFNA2, IGSF3, lkB-Tp53, KLF4, LAPTM4B, MT1L, NFKBIA, PQLC3, PROM1, PYHIN1 (includes EG:149628), SCN3B, SLC19A2, SOD2, SPHK2, SP0N1, TACC3, TBX3, TEP1, TERT, TMC1, TP53, TRIM14, TRIM22, TRIM28, Ube3, ZNF10 | 19 | 13 | Cellular Growth and Proliferation, Cell Cycle, Cell Death |
| 7 | ABCC4, ACCN1, ACCN2, ATXN1, BCAS1, beta-estradiol, BICD1, CACNB1, DDX54, GMEB2, GRM2, GRM3, HSD17B12, HUS1B, IFT122, MAL, MATN2, MIR133A-1, NAPB, NARS, NR1I3, NR3C1, NSF, OBSCN (includes EG:84033), PAPSS2, PICK1, PTP4A2, RAB6A, SAPS2, SEPT3, SLC1A6, SULT1A1, TGTP1, ZCCHC2, ZNF804A | 18 | 12 | Cancer, Psychological Disorders, Cell-To-Cell Signaling and Interaction |
| 8 | 4930444G20RIK, ADAMTS14, alcohol group acceptor phosphotransferase, amino acids, ASTL, CPA5, DMPK, DPEP3, FAM70A, GRAMD1B, HIPK1, IMMP2L, KIAA2022, LTK, MIR129-2 (includes EG:406918), MIR195 (includes EG:406971), MIR362 (includes EG:574030), MMP1B, NAALADL1, PAK7, PEPC (includes EG:109616), peptidase, PRKX, PRPF4B, PRT5, PRT6, PTPRD, PTPRM, RNF150, SENP5 (includes EG:303874), SOLH, TMEM132E, TMPRSS11D, UPG2, YME1L1 | 18 | 12 | Genetic Disorder, Skeletal and Muscular Disorders, Protein Degradation |
| 9 | ABLIM, Akt, ATP9A, CREBL2, DHCR7, DHRS3, DYRK3, FASN, FSH, GATA3, GK7P, IFN Beta, IgG, IGKV1-117, IKBKB, IL12 (complex), Insulin, Interferon alpha, Jnk, Jun-ATF2, Lh, LOC81691, MAS1, P38 MAPK, PI4K2A, Pka, QRFP, RASAL2,RPA1, TAPBP, TFRC, TP53I11, TRIL, USP18, ZNF668 ACOT2, ACOT4, ACOT5, ACOT7, ACOT8, ACOT9, ACOT1 (includes EG:26897), ACOT1 (includes EG:641371), BAAT, C22ORF28, C2ORF47, C3ORF26, CLEC12B, FASTKD2, GOLIM4, GPX7, GSTK1, HNF4A, LAS1L, MTR, OGFR, palmitoyl-CoA hydrolase | 16 | 11 | Cellular Response to Therapeutics, Lipid Metabolism, Reproductive System Development and Function |
| 10 | PPT1, PTPN11, SEL1L3, TCF19, TDRKH, TLN1, TMEM63A, T0R1AIP2, TSC22D1, USMG5, UTP11L (includes EG:51118), VEPH1, VN1R1 | 16 | 11 | Lipid Metabolism, Nucleic Acid Metabolism, Small Molecule Biochemistry |
| 11 | ADAMTS3, ADAMTS13, ADK, AKT1, Akt-Calmodulin-Hsp9O-Nos3, ATG9B, ATP13A3 (includes EG:79572), C1QC, CACHD1, CASP3, COL6A1, CORO1C, cyclic AMP, F2, FGL2, FXYD5, Lamin, L0XL2, LPHN1, MAGI2, N0S3, PDE11A, PDE1A, PDE4C, PDE7A, PLXNAl, PPT1, PTGDS, SLC12A7, SOLH, SOX15, TGFB1, USP25, WNK3, ZMIZ1 | 16 | 11 | Reproductive System Disease, Cell Morphology, Inflammatory Disease |
| 12 | AGRN, ARPP19, B9D1, CEBPB, CENPV, CLPX, COPG2, Creb, CXCR6 (includes EG:10663), DDX42, DRD1, EN03, ENTPD2, GPR183, HSPD1, HTT, KLF16, KPTN, LDB2, LMO4, MYL4, NDUFA3, PCTP, PSG4, RLIM, SCHIP1, SEPP1, SF3A2, SMARCA4, SRGAP3, SRRT, TNNI2, TRAP1, VIPR2, ZNF675 | 14 | 10 | Carbohydrate Metabolism, Cell Signaling, Nucleic Acid Metabolism |
The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (in bold) are genes identified in our list of differentially expressed genes. Networks shown here are those with network scores > 3.0.
Figure 3.
Top network overrepresented by differentially expressed genes. The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (shaded) are genes identified in our list of differentially expressed genes.
In the qRT-PCR experiment to confirm microarray-based measurements conducted on selected differentially expressed genes (of CLEC family of genes or functionally related genes), similar direction of fold change differences (and for some, similar size of fold change differences) between preeclampsia cases and controls were observed for most genes (6/8, 75%) (Table 5). However, most of the p-values in Student's T-test comparisons were not statistically significant.
Table 5.
Microarray and qRT-PCR expression measurement comparisons
| Gene symbol | Gene description | qRT-PCR | Microarray | ||
|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | ||
| CLEC11A | C-type lectin domain family 11, member A | 1.06 | 0.763 | -1.56 | 0.006 |
| CLEC12A | C-type lectin domain family 12, member A | -1.52 | 0.22 | -1.54 | 0.018 |
| CLEC12B | C-type lectin domain family 12, member B | -2.08 | 0.024 | -1.59 | 0.001 |
| MMP25 | Matrix metallopeptidase 25 | 1.09 | 0.705 | 1.95 | 0.019 |
| FGFR1 | Fibroblast growth factor receptor 1 | -1.24 | 0.247 | -1.52 | 0.013 |
| LTK | Leukocyte receptor tyrosine kinase | -1.25 | 0.428 | -1.96 | 0.027 |
| PML | Promyelocytic leukemia protein | -1.08 | 0.685 | -1.86 | 0.021 |
| PPP1R13L | Protein phosphatase 1, regulatory subunit 13 like | 1.05 | 0.813 | -1.51 | 0.013 |
In the promoter analysis of common regulatory sequences (motifs) of differentially expressed genes, binding sites of transcription factors Sp1 (specificity protein 1), MAZ (MYC associated zinc finger protein) and MZF1 (myeloid zinc finger 1) were identified (Figure 4).
Figure 4.
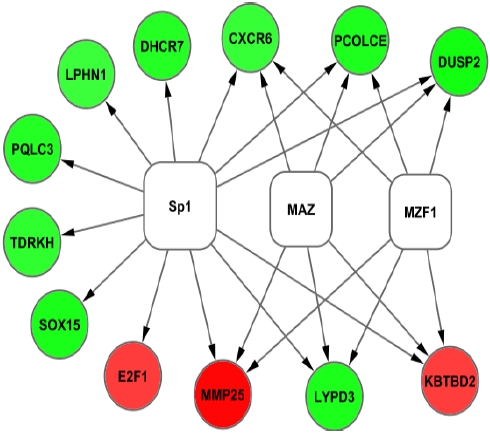
Promoter analysis results of differentially expressed genes. Inferred network of differentially expressed genes (Red = up regulated and Green=down regulated) in preeclampsia and transcription factors (White). Transcription factors were identified by their binding to over expressed promoter sequences in the differentially expressed genes.
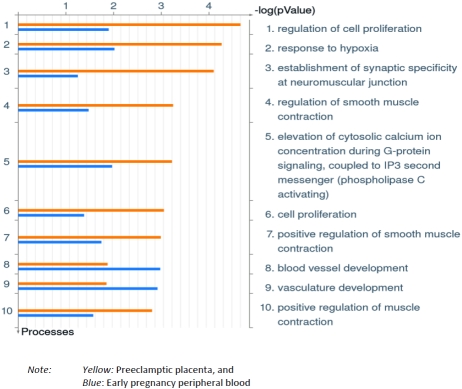
Results of GO comparisons of preeclampsia related differentially expressed genes in the current experiment with preeclampsia related differentially expressed genes in placenta at-delivery, reported before [7], are presented in Figure 5. GO processes of cell proliferation, response to hypoxia and smooth muscle contraction were over represented in preeclamptic placenta while GO processes of vasculature (blood vessel) development were over represented in early pregnancy peripheral blood among women who later developed preeclampsia.
Figure 5.
Comparison of gene ontology processes. Gene ontology (GO) processes over represented by differentially expressed genes in early pregnancy maternal peripheral blood (blue) and preeclamptic placenta (yellow).
Discussion
We demonstrated that preeclampsia is associated with differential gene expression in early pregnancy maternal peripheral blood. Genes participating in abnormal placentation (e.g COL1A1), immune/inflammation response (e.g. IKBKB) and cellular development (including cell cycle) (e.g. RBI) were differentially expressed. We identified transcription factors (e.g. Spl, MAZ and MZF1) that may account for co-expression of differentially expressed genes. Comparison of preeclampsia related gene expression profiles of early pregnancy peripheral blood and placenta (at-delivery) suggest gestational age and tissue specific differences in pathophysiological processes (vasculature development versus hypoxia response, respectively) involved in preeclampsia.
Previous studies have investigated peripheral blood gene expression in relation to preeclampsia [10, 12, 18-20]. Okazaki et al reported up-regulation of pregnancy specific beta-1 glycoprotein and trophoblast glycoprotein in peripheral blood of women with preeclampsia at 38-39 weeks of gestation [18]. Purwosunu et al, in a qRT-PCR based study of peripheral blood samples collected around 39 weeks of gestation, reported that expression of CRH, PLAC1 and P-Selectin were up-regulated in women with preeclampsia [19]. In another qRT-PCR based study, Purwosunu and colleagues have reported differential regulation of angiogenesis-related genes including Flt-1 and VEGF in peripheral blood of women with preeclampsia at 38-39 weeks of gestation [20]. Sun et al reported that 72 genes involved in cell proliferation, smooth muscle contraction and immune response were differentially expressed in peripheral blood of women with preeclampsia at 24-32 weeks of gestation [21]. In a follow-up study of chorionic villus gene expression study in early pregnancy (11 weeks), Farina etal investigated expressions of selected genes in third trimester peripheral blood of women who developed preeclampsia [10]. They reported up regulation of ADD1, BTD7, CLDN6, LTF and MAS1 in third trimester peripheral blood of women with preeclampsia. Recently, Sekizawa et al investigated expressions of selected candidate genes in peripheral blood at 16-17 weeks to identify early pregnancy markers of preeclampsia [12]. In their study, expressions of FLT-1, ENG, P-Selectin, PLAC1, P1GF and HO-1 were deregulated in the case group while no differences were present between cases and controls in expressions of TGFB1, VEGF and S0D[12].
Investigators have also studied gene expression profiles in chorionic villus tissue samples collected in early pregnancy in relation to risk of preeclampsia [10-11]. Farina et al reported preeclampsia related differential expression of genes involved in trophoblast invasion, inflammation, endothelial dysfunction, angiogenesis and blood pressure control in chorionic villus samples in early pregnancy (11 weeks) [10]. Similarly, Founds et al reported deregulation of genes related to inflammation/immune regulation in early pregnancy (10-12 weeks) chorionic villus samples in women who later developed preeclampsia [11]. Genes involved in hypoxia or oxidative stress responses were not differentially expressed in their samples, similar to our findings. In sum, review of current and previous preeclampsia related gene expression profiles from blood and placental tissues suggest early pregnancy changes consistent with alterations in angiogenesis and immune/inflammatory response in contrast to late pregnancy changes which are consistent with alterations in response to hy-poxemia or oxidative stress and subsequent endothelial dysfunction.
In our study, several genes that participate in abnormal placentation were differentially expressed in preeclampsia. In their candidate gene study, Goddard et a I reported associations of variations in the COL1A1 gene with risk of preeclampsia [22]. COL1A1 is a gene coding for a protein in collagen metabolism (similar to COL5A3, also differentially expressed in our study) which influence maternal extracellular matrix composition and subsequently trophoblast migration [23]. NRTK2 is a brain derived neurotrophin family of proteins known to activate the high-affinity tyrosine kinase [24]. Kawamura et al, using in vitro and in vivo studies, have previously demonstrated important roles of the tyrosine kinase B signaling system and related neurotrophins in implantation and placental development through regulation of trophoblast cell growth [24].
Several genes in the immune response/ inflammation and cell cycle pathways were also differentially expressed related to preeclampsia in our study. For instance, genes constituting the CLEC family of genes (e.g. CLEC11A, CLEC12A and CLEC12B) were down regulated. These C-type lectin receptors play crucial roles in immunity and homeostasis, particularly in pathogen and self-antigen recognition, pathomechanisms that have been implicated in preeclampsia [25-27]. Regulatory signal pathways of the inflammatory system involving TNFRSFlATRAFs, IKBKB and NFKB genes have been described [28]. IKBKB was differentially expressed in our study, while NFKB plays a central role in the top network that was over represented by differentially expressed genes. Genes participating in the RB_E2R1 cell cycle pathway were also differentially expressed in our study. While most research in this pathway has been done in cancer research, recently, interest in this pathway related to vascular disorders has increased following identification of E2F1 binding sites in promoters of angiogenesis related genes (e.g. FLT-1) [29].
We identified putative transcription factors (i.e., Spl, MAZ and MZF1) that may be responsible for co-expression of differentially expressed genes. Sp1 has been associated with transcription of genes involved in syncytiotrphoblast differentiation such as the PSG family of genes (e.g. PSG4 up regulated in our study), endoglin and TGFpi and 2 other genes [30]. Further research in this area may enhance understanding of mechanisms of abnormal syncytiotrophoblast differentiation and related pathologies such as preeclampsia.
Our study has several strengths and limitations. It is the first global microarray based study investigating risk of preeclampsia and early pregnancy differential gene expression in peripheral blood, to our knowledge. Evaluation of functions and functional relationships of differentially expressed genes, for example using GO processes, as observed in past reports, enhances comparison of findings across studies [31]. By comparing preeclampsia related differential gene expression in early pregnancy peripheral blood and placenta at-delivery, we were able to present corroborative evidence for recent hypotheses that seek to elucidate gestational age and/or tissue specific gene expression changes associated with preeclampsia [4].
Several limitations of our study deserve mention. Single measurement of peripheral blood gene expression may not provide a full picture of gene expression changes across gestation. Evaluation of whole blood gene expression, a potentially heterogeneous cell population, does not allow comparisons of expression differences across similar cell subtypes. We were able to confirm microarray-based measurement for approximately 75% of genes in our confirmatory qRT-PCR study. This is comparable to other previous reports that range between 60-75% [32-33]. Further, most fold change differences observed were in the same direction in both experiments. For the two genes with different fold change directions (up or down regulation) between the two experiments, the qRT-PCR based gene expression differences were close to 1 (1.05 for PPP1R13L and 1.06 for CLEC11A).
In summary, we demonstrated maternal early pregnancy peripheral blood gene expression in early pregnancy. Differentially expressed genes participate in cellular processes of placentation, immune function/inflammation and cell growth (cell cycle). Besides improving understanding of pathogenesis of preeclampsia, early pregnancy peripheral blood gene expression profiling may provide critical windows of opportunity for disease prevention, early detection and/or treatment.
Acknowledgments
The authors are indebted to the participants of the Omega study for their cooperation. They are also grateful for the technical expertise of staff of the Center for Perinatal Studies, Swedish Medical Center. This work was supported by grants from the National Institute of Child Health and Human Development, National Institutes of Health (HD/HL R01-32562 and R01-055566).
References
- 1.Baumwell S, Karumanchi SA. Pre-Eclampsia: Clinical Manifestations and Molecular Mechanisms. NephronClin Pract. 2007;106:c72–c81. doi: 10.1159/000101801. [DOI] [PubMed] [Google Scholar]
- 2.Mutter WP, Karumanchi SA. Molecular mechanisms of preeclampsia. Microvasc Res. 2008;75:1–8. doi: 10.1016/j.mvr.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanasaki K, Kalluri R. The biology of preeclampsia. Kidney Int. 2009;76:831–837. doi: 10.1038/ki.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foidart JM, Schaaps JP, Chantraine F, Munaut C, Lorquet S. Dysregulation of anti-angiogenic agents (sFlt-1, PLGF, and sEndoglin) in preeclampsia—a step forward but not the definitive answer. J Reprod Immunol. 2009;82:106–111. doi: 10.1016/j.jri.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Chappell S, Morgan L. Searching for genetic clues to the causes of preeclampsia. Clin Sci (London) 2006;110:443–458. doi: 10.1042/CS20050323. [DOI] [PubMed] [Google Scholar]
- 6.Sitras V, Paulssen RH, Grønaas H, Leirvik J, Hanssen TA, Vartun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–433. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J ObstetGynecol. 2008;199(566):e1–11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe preeclampsia. Placenta. 2007;28:487–497. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Reimer T, Koczan D, Gerber B, Richter D, Thiesen HJ, Friese K. Microarray analysis of differentially expressed genes in placental tissue of preeclampsia: up-regulation of obesity-related genes. Mol Hum Reprod. 2002;8:674–680. doi: 10.1093/molehr/8.7.674. [DOI] [PubMed] [Google Scholar]
- 10.Farina A, Morano D, Arcelli D, De Sanctis P, Sekizawa A, Purwosunu Y, Zucchini C, Simonazzi G, Okai T, Rizzo N. Gene expression in chorionic villous samples at 11 weeks of gestation in women who develop preeclampsia later in pregnancy: implications for screening. Prenat Diagn. 2009;29:1038–1044. doi: 10.1002/pd.2344. [DOI] [PubMed] [Google Scholar]
- 11.Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30:15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekizawa A, Purwosunu Y, Farina A, Shimizu H, Nakamura M, Wibowo N, Rizzo N, Okai T. Prediction of preeclampsia by an analysis of placenta-derived cellular mRNA in the blood of pregnant women at 15-20 weeks of gestation. BJOG. 2010;117:557–564. doi: 10.1111/j.1471-0528.2010.02491.x. [DOI] [PubMed] [Google Scholar]
- 13.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 14.Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 15.Yeung KY, Ruzzo WL. Prinicipal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooghe B, Hulpiau P, van Roy F, De Bleser P. ConTra: a promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res. 2008;36:W128–W132. doi: 10.1093/nar/gkn195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okazaki S, Sekizawa A, Purwosunu Y, Farina A, Wibowo N, Okai T. Placenta-derived, cellular messenger RNA expression in the maternal blood of preeclamptic women. Obstet Gynecol. 2007;110:1130–1136. doi: 10.1097/01.AOG.0000286761.11436.67. [DOI] [PubMed] [Google Scholar]
- 19.Purwosunu Y, Sekizawa A, Farina A, Wibowo N, Okazaki S, Nakamura M, Samura O, Fujito N, Okai T. Cell-free mRNA concentrations of CRH, PLAC1, and selectin-P are increased in the plasma of pregnant women with preeclampsia. Prenat Diagn. 2007;27:772–777. doi: 10.1002/pd.1780. [DOI] [PubMed] [Google Scholar]
- 20.Purwosunu Y, Sekizawa A, Yoshimura S, Farina A, Wibowo N, Nakamura M, Shimizu H, Okai T. Expression of angiogenesis-related genes in the cellular component of the blood of preeclamptic women. Reprod Sci. 2009;16:857–864. doi: 10.1177/1933719109336622. [DOI] [PubMed] [Google Scholar]
- 21.Sun CJ, Zhang L, Zhang WY. Gene expression profiling of maternal blood in early onset severe preeclampsia: identification of novel biomarkers. J Perinat Med. 2009;37:609–616. doi: 10.1515/JPM.2009.103. [DOI] [PubMed] [Google Scholar]
- 22.Goddard KA, Tromp G, Romero R, Olson JM, Lu Q, Xu Z, Parimi N, Nien JK, Gomez R, Behnke E, Solari M, Espinoza J, Santolaya J, Chaiworapongsa T, Lenk GM, Volkenant K, Anant MK, Salisbury BA, Carr J, Lee MS, Vovis GF, Kuivaniemi H. Candidate-gene association study of mothers with preeclampsia, and their infants, analyzing 775 SNPs in 190 genes. Hum Hered. 2007;63:1–16. doi: 10.1159/000097926. [DOI] [PubMed] [Google Scholar]
- 23.Hess AP, Hamilton AE, Talbi S, Dosiou C, Nyegaard M, Nayak N, Genbecev-Krtolica O, Mavrogianis P, Ferrer K, Kruessel J, Fazleabas AT, Fisher SJ, Giudice LC. Decidual stromal cell response to paracrine signals from the trophoblast: amplification of immune and angiogenic modulators. Biol Reprod. 2007;76:102–117. doi: 10.1095/biolreprod.106.054791. [DOI] [PubMed] [Google Scholar]
- 24.Kawamura K, Kawamura N, Sato W, Fukuda J, Kumagai J, Tanaka T. Brain-derived neurotrophic factor promotes implantation and subsequent placental development by stimulating trophoblast cell growth and survival. Endocrinology. 2009;150:3774–3782. doi: 10.1210/en.2009-0213. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann SC, Schellack C, Textor S, Konold S, Schmitz D, Cerwenka A, Pflanza S, Watzl C. Identification of CLEC12B, an inhibitory receptor on myeloid cells. J Biol Chem. 2007;282:22370–22375. doi: 10.1074/jbc.M704250200. [DOI] [PubMed] [Google Scholar]
- 26.Thebault P, Lhermite N, Tilly G, Le Texier L, Quillard T, Heslan M, Anegon I, Soulillou JP, Brouard S, Charreau B, Cuturi MC, Chiffoleau E. The C-type lectin-like receptor CLEC-1, expressed by myeloid cells and endothelial cells, is up-regulated by immunoregulatory mediators and moderates T cell activation. J Immunol. 2009;183:3099–3108. doi: 10.4049/jimmunol.0803767. [DOI] [PubMed] [Google Scholar]
- 27.Bonney EA. Preeclampsia: a view through the danger model. J Reprod Immunol. 2007;76:68–74. doi: 10.1016/j.jri.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Li H, Bocking AD, Challis JR. Tumor Necrosis Factor Stimulates Matrix Metalloproteinase 9 Secretion from Cultured Human Chorionic Trophoblast Cells Through TNF Receptor 1 Signaling to IKBKB-NFKB and MAPK1/3 Pathway. Biol Reprod. 2010 doi: 10.1095/biolreprod.109.082578. May 12. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Pillai S, Kovacs M, Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70:4931–4940. doi: 10.1158/0008-5472.CAN-10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nores R, Blanchon L, López-Díaz F, Bocco JL, Patrito LC, Sapin V, Panzetta-Dutari GM. Transcriptional control of the human pregnancy-specific glycoprotein 5 gene is dependent on two GT-boxes recognized by the ubiquitous specificity protein 1 (Sp1) transcription factor. Placenta. 2004;25:9–19. doi: 10.1016/S0143-4004(03)00213-3. [DOI] [PubMed] [Google Scholar]
- 31.Toxicogenomics Research Consortium. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 32.Jenson SD, Robetorye RS, Bohling SD, Schumacher JA, Morgan JW, Lim MS, Elenitoba-Johnson KS. Validation of cDNA microarray gene expression data obtained from linearly amplified RNA. Mol Pathol. 2003;56:307–312. doi: 10.1136/mp.56.6.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]