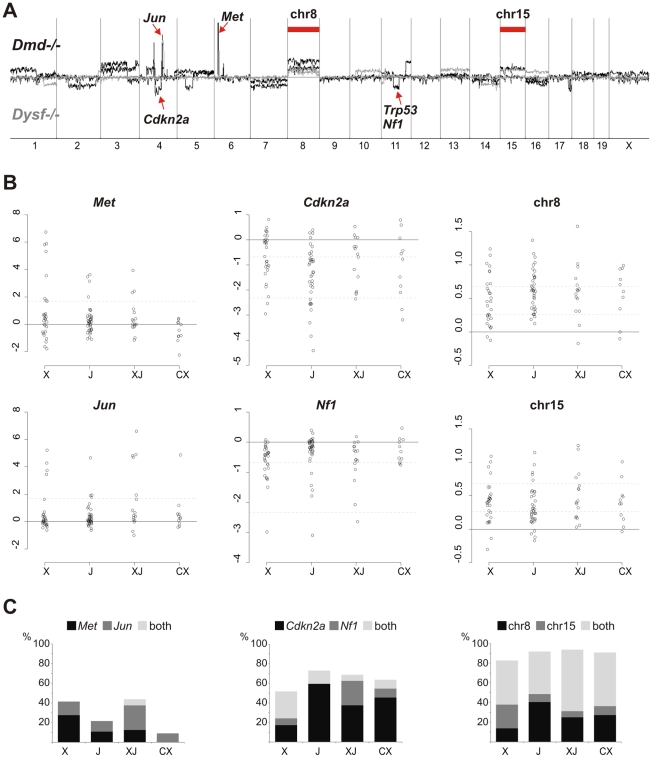
Figure 2. Common non-random genetic lesions in MD-mouse sarcomas.
(A) Chromosomal aberration profiles from Agilent aCGH experiments performed on tumor DNA isolated from sarcomas of n = 4 Dmd −/− (black) and n = 4 Dysf −/− (grey) mice revealing gross genomic instability, specific losses at the Cdkn2a, Nf1, and Trp53 tumor suppressor loci, amplification of Met and Jun oncogenes, and recurrent gains of whole chromosome 8 and 15. (B) Real-time PCR quantification of Met and Jun oncogene amplification, losses at the Cdkn2a and Nf1 loci, and of chromosome 8 and 15 copy numbers in n = 98 sarcomas from Dmd −/− (X), Dysf −/− (J), Dmd −/− Dysf −/− (XJ), and Dmd −/− Capn3 −/− (CX) mice. Relative copy numbers (shown as log2) were calculated by the ΔΔCt-method. Dashed lines indicate log2-thresholds that were set under the assumption of 80% tumor cell content: gene amplification (4-fold; Met and Jun), deletions (first line for heterozygous losses, 0.5-fold, second line for homozygous losses; Cdkn2a and Nf1), and chromosome copy numbers (3 and 4). (C) Frequency plots for Met and Jun amplification, Cdkn2a and Nf1 deletions, and chromosome 8/15 gains from the n = 98 sarcomas shown in (B).