Abstract
A new species with remarkable morphology, Nectria eustromatica, is described, based on morphology of the teleomorph and anamorph, ecology and molecular phylogenetic analyses. Nectria eustromatica is characterized by sphaeroid perithecia immersed in pseudoparenchymatous stromata formed singly or collectively on a subiculum. Despite its deviating teleomorph morphology, it is placed within Nectria sensu stricto in phylogenetic analyses of a combined dataset of LSU, ITS, rpb2 and tef1 sequences with high internal support. Nectria eustromatica has been collected specifically on Hippocrepis (Coronilla) emerus in southern Europe. The anamorph of N. eustromatica shares morphological traits with the genera Stilbella and Tubercularia but produces non-phialidic macroconidia in addition to phialoconidia.
Keywords: Ascomycetes, Hypocrea, ITS, LSU, morphology, Nectria, phylogenetic markers, rpb2, sequence analysis, Stilbella, Stilbocrea, tef1, Tubercularia
INTRODUCTION
Ascomata and stromata of the Hypocreales have usually light or bright colors (Rossman et al. 1999). Exceptions for example are Hypocrea lixii Pat. (Jaklitsch 2009) or H. schweinitzii (Fr.) Sacc. and similar species (Samuels et al. 1998) of the Hypocreaceae. In the Nectriaceae the stroma when present is typically a hypostroma, a loose or compact, pros- or pseudoparenchymatous layer or pillow, which gives rise to more or less free, superficial perithecia on its top, usually with clearly discernible perithecial contours, even when tightly associated in clusters. In genera of the Bionectriaceae such as Stilbocrea Pat. (Rossman et al. 1999) ascomata are immersed in light-colored, prosenchymatous stromata.
Intense searches for Hypocrea teleomorphs in Europe have revealed a fungus that superficially resembles representatives of the Hypocreaceae or other stromatic ascomycetes because of its dark brown to nearly black stromata. However the centrum morphology of this fungus is nectriaceous. This fungus is described here as a new species of Nectria (Fr.) Fr.
MATERIALS AND METHODS
Isolates and specimens
Taxon names and accession numbers of gene sequences included in this study are provided (Table I); data on isolates sequenced in the present study also are provided (Table II). Representative isolates have been deposited at the Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands (CBS). Specimens have been deposited in the Herbarium of the Institute of Botany, University of Vienna (WU).
Table I.
GenBank accession numbers of the sequences used for multigene phylogenetic analyses. Sequences starting with HM were generated in the present study
| Taxon | Strain | LSU | ITS | rpb2 | tef1 |
|---|---|---|---|---|---|
|
Bionectria ochroleuca (Schwein.) Schroers & Samuels |
CCFC 226708 CBS 376.55 AFTOL-ID 187 |
AY283558 — — |
— AF358239 — |
— — DQ862013 |
— — DQ862029 |
| Cosmospora coccinea Rabenh. | A.R. 2741 CBS 114050 |
AY489734 — |
— FJ474072 |
— DQ522438 |
AY489629 — |
|
Cosmospora episphaeria (Tode) Rossman & Samuels |
G.J.S. 98-160 G.J.S. 88-29 |
— AY015625 |
FJ474073 — |
—
— |
—
— |
|
Cosmospora vilior (Starbäck) Rossman & Samuels |
Guardbridge 20 G.J.S. 96-186 |
— AY015626 |
GU726755 — |
—
— |
—
— |
|
Haematonectria haematococca (Berk. & Broome) Samuels & Rossman |
voucher 83364 not specified ATCC MYA-4622 |
DQ119558 — |
GU327638 |
Genomea — |
Genomea — |
|
Hydropisphaera peziza (Tode) Dumort. |
G.J.S. 92-101 CBS 102038 |
AY489730 — |
—
— |
— DQ522444 |
AY489625 — |
| Hypocrea rufa (Pers.) Fr. | CBS 114374 (G.J.S. 89-127) C.P.K. 1998 |
AY489726 — |
— DQ677656 |
EF692510 — |
— DQ672616 |
| Nectria aquifolii (Fr.) Berk. | CBS 127381 | HM534891 | HM534891 | HM534881 | HM534870 |
|
Nectria aurantiaca (Tul. & C. Tul.) Jacz. |
CBS 236.29 | HM534892 | HM534892 | HM534882 | HM534871 |
| Nectria berolinensis (Sacc.) Cooke | CBS 127382 | HM534893 | HM534893 | HM534883 | HM534872 |
| Nectria cinnabarina (Tode) Fr. | CBS 127383 | HM534894 | HM534894 | HM534884 | HM534873 |
| Nectria coryli Fuckel | CBS 127384 | HM534895 | HM534895 | HM534885 | HM534874 |
|
Nectria eustromatica Jaklitsch & Voglmayr |
CBS 121896 (NC) | HM534896 | HM534896 | HM534886 | HM534875 |
| Nectria eustromatica | CBS 125578 (NC1) | HM534897 | HM534897 | HM534887 | HM534876 |
| Nectria lamyi (Desm.) De Not. | CBS 127385 | HM534898 | HM534898 | HM534888 | HM534877 |
|
Nectria pseudotrichia (Schwein.) Berk. & M.A. Curtis |
CBS 641.83 | HM534899 | HM534899 | HM534889 | HM534878 |
| Nectria sinopica (Fr.) Fr. | CBS 127386 | HM534900 | HM534900 | HM534890 | HM534879 |
|
Neonectria coccinea (Pers.) Rossman & Samuels |
CBS 237.29 CBS 29181 CBS 119159 CBS 118914 |
AY677327 — — — |
— FJ474075 — — |
— — DQ789819 — |
— — — DQ789688 |
|
Neonectria ditissima (Tul. & C. Tul.) Samuels & Rossman |
CBS 226.31 CBS 117752 G.J.S. 94-12 CBS 118927 |
AY677330 — — — |
— DQ178168 — — |
— — DQ789823 — |
— — — DQ789743 |
|
Neonectria punicea (J.C. Schmidt) Castl. & Rossman |
CBS 124262 CBS 119724 |
HM534901 — |
HM534901 — |
— DQ789753 |
HM534880 — |
|
Pseudonectria rousseliana (Mont.) Wollenw. |
A.R. 2716 not specified CBS 114049 |
U17416 — — |
— FJ555527 — |
— — DQ522459 |
AF543780 — — |
|
Roumegueriella rufula (Berk. & Broome) Malloch & Cain |
G.J.S. 91-164 | EF469082 | — | EF469116 | EF469070 |
|
Sphaerostilbella aureonitens (Tul. & C. Tul.) Seifert, Samuels & W. Gams |
TFC 96-77 G.J.S. 74-87 |
AF160246 |
FJ442633 |
FJ442763 |
DQ834452 |
Retrieved from the JGI database (http://genome.jgi-psf.org/).
Table II.
Source data, CBS culture numbers and herbarium vouchers of the specimens sequenced in the present study
| Taxon | Geographic origin, year, collector | Host | Strain | Herbarium voucher |
|---|---|---|---|---|
| Nectria aquifolii | UK, Surrey, Royal Botanic Gardens Kew, 11 Nov 2008, H. Voglmayr |
Ilex aquifolium | CBS 127381 | WU 30360 |
| Nectria aurantiaca | UK, Bristol, Oct 1929, E.W. Mason | Ulmus campestris | CBS 236.29 | — |
| Nectria berolinensis | Austria, Wien, Floridsdorf, 13 Apr 2009, W. Jaklitsch |
Ribes sanguinea | CBS 127382 | WU 30361 |
| Nectria cinnabarina | Austria, Niederösterreich, Litschau, 14 Sep 2009, W. Jaklitsch |
Frangula alnus | CBS 127383 | — |
| Nectria coryli | Austria, Oberösterreich, St. Willibald, 22 May 2009, H. Voglmayr |
Pyrus communis | CBS 127384 | WU 30362 |
| Nectria eustromatica | Croatia, Opatija, Mošcenička Draga, 29 Mar 2007, W. Jaklitsch & H. Voglmayr |
Hippocrepis emerus | CBS 121896 (NC) | WU 30194 |
| Nectria eustromatica | Italy, Lazio, Bagnaia, Prov. Viterbo, 28 Jul 2009, W. Jaklitsch & H. Voglmayr |
Hippocrepis emerus | CBS 125578 (NC1) |
WU 30195 |
| Nectria lamyi | Austria, Wien, Floridsdorf, 31 May 2009, W. Jaklitsch |
Berberis thunbergii | CBS 127385 | WU 30363 |
| Nectria pseudotrichia | Venezuela; Edo Tachira, near La Fria, 21 Jul 1971, K.P. Dumont & al. |
unidentified wood | CBS 641.83 | — |
| Nectria sinopica | Austria, Niederösterreich, Maierhö fen, 20 Jun 2009, W. Jaklitsch & H. Voglmayr |
Hedera helix | CBS 127386 | WU 30364 |
| Neonectria punicea | Austria, Kärnten, St. Margareten i. Rosental, 2 Nov 2008, W. Jaklitsch |
Frangula alnus | CBS 124262 | WU 30365 |
Ascospore isolates were prepared as described by Jaklitsch (2009). Cultures were grown in 9 cm diam Petri dishes either in the dark at 15 C, in daylight or with alternating 12 h cool white fluorescent light and 12 h darkness at 20–25 C on oatmeal agar (OA, Sigma), 2% malt extract agar (MEA), potato dextrose agar (PDA), low nutrient agar (SNA) and cornmeal dextrose agar (CMD, Jaklitsch 2009).
Morphological observations
Conidiation structures were examined, measured and photographed on a compound microscope from cultures grown on SNA, PDA, OA or MEA after mounting in 3% KOH. Dry stromata were rehydrated overnight with water vapor in a closed glass chamber at room temperature, treated briefly with 3% KOH, embedded in Tissue-Tek O.C.T. Compound 4583 (Sakura Finetek Europe B.V., Zoeterwoude, the Netherlands) and sectioned 10 μm thick with a freezing microtome. Sections were measured and photographed in lactic acid or in 50% glycerol or 3% KOH where noted. Asci and ascospores were measured in separate preparations in 3% KOH. (See Jaklitsch [2009] for the terminology of stromatal traits.) Measurements are reported as maxima and minima in parentheses and the mean plus and minus the standard deviation of a number of measurements given in parentheses. Nomarski differential interference contrast (DIC) was used for observations and measurements. Images were recorded with the Nikon Coolpix 4500 or DS-U2 digital cameras. Measurements were made with NIS-Elements D 3.0 software.
DNA extraction, PCR amplifications and sequencing
Mycelium for DNA extraction was grown in liquid malt extract culture, harvested, freeze-dried and ground according to Voglmayr and Jaklitsch (2008). Genomic DNA was extracted with the modified CTAB method described in Riethmüller et al. (2002). A 1.6 kb fragment containing partial SSU, ITS1, 5.8S, ITS2 and partial LSU was amplified with the primer pair V9G (de Hoog and Gerrits van den Ende 1998) and LR5 (Vilgalys and Hester 1990). A 1.1 kb fragment of RNA Polymerase II subunit B (rpb2) was amplified with the primer pair fRPB2-5f and fRPB2-7cr (Liu et al. 1999). A 1.3 kb fragment of the tef1 gene encoding translation elongation factor 1 alpha was amplified with the primer pair EF1728F (Chaverri and Samuels 2003) and TEF1LLErev (Jaklitsch et al. 2005). This fragment includes the fourth and the fifth intron and a part of the last large exon. PCR products were purified by an enzymatic PCR cleanup (Werle et al. 1994) as described in Voglmayr and Jaklitsch (2008). DNA was cycle sequenced with the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit 3.1 (Applied Biosystems, Warrington, UK) and an automated DNA sequencer (ABI 3730xl Genetic Analyzer, Applied Biosystems) with the same primers as in PCR; in addition for the SSU-ITS-LSU fragment primers LR3 (Vilgalys and Hester 1990), ITS4 (White et al. 1990) and F5.8Sr (5′TGCGTTCAAARATTCGATG 3′) were used as internal sequencing primers. Due to abrupt signal loss in some Nectria species with GC-rich sequence regions in the ITS, it sometimes was necessary to use F5.8Sf (5′CAACAACGGATCTCTTGGYTC 3′) and ITS5 (White et al. 1990) as additional internal sequencing primers. (All sequences generated in this study are listed in Table I.)
Molecular phylogenetic analyses
For the phylogenetic analyses representative LSU, ITS, tef1 and rpb2 sequences of Hypocreales were selected from GenBank according to a BLAST query that revealed a high sequence homology of LSU sequences of the new species to Nectria sensu stricto (Table I). However because only few sequences were available for Nectria the dataset was complemented with some representative Nectria species collected by the authors or obtained from CBS (Table II). The final matrix contained sequences from 23 taxa, including Hypocrea rufa and Sphaerostilbella aureonitens as outgroups. All alignments were produced with Muscle 3.6 (Edgar 2004). A combined dataset of LSU, ITS, rpb2 and tef1 sequences was used for the analyses. After the exclusion of leading and trailing gap regions and of ambiguously aligned positions in the ITS and tef1 alignments, the combined matrix contained 3645 characters (viz. 834 nucleotides from the LSU, 563 from the ITS1-5.8S-ITS2 region, 1110 nucleotides from rpb2 and 1138 nucleotides from tef1). Incongruence between the different gene regions included in the multigene analyses was evaluated by comparison of maximum parsimony (MP) bootstrap trees of the individual gene regions, which were calculated with the same parameters as for the combined analysis given below. Incongruence receiving MP bootstrap support under 70% was considered low, posing no major obstacle for the combined analyses. Maximum parsimony (MP) analyses were performed with PAUP* 4.0 b10 (Swofford 2002) with 1000 replicates of heuristic search with random addition of sequences and subsequent tbr branch swapping (multrees option in effect, collapse = maxbrlen, steepest descent option not in effect). All molecular characters were unordered and given equal weight. Analyses were performed with gaps treated as missing data. Bootstrap analysis with 1000 replicates was performed in the same way but using 10 rounds of random sequence addition and subsequent tbr branch swapping during each bootstrap replicate. For maximum likelihood (ML) and Bayesian analyses first the appropriate models of sequence substitution were selected with Modeltest 3.6 (Posada and Crandall 1998) with the Akaike information criterion (AIC). These nucleotide substitution models were revealed by Modeltest: for rpb2 the general time reversible model was chosen, additionally assuming a proportion of invariant sites with gamma-distributed substitution rates of the remaining sites (GTR + I + G); for LSU the ITS and tef1 matrices the model of Tamura and Nei (1993) was selected, additionally assuming a proportion of invariant sites with gamma-distributed substitution rates of the remaining sites (TRN + I + G). Because the latter model could not be implemented in ML and Bayesian analyses the GTR + I + G model was used for all sequence regions of the combined matrix. For the analyses partitioned substitution models were implemented for each gene. For ML analyses 200 rounds of random addition of sequences as well as 200 bootstrap replicates were computed with RAxML 7.0.4 (Stamatakis 2006) with the GTRMIXI and GTRCAT algorithms respectively. GTRCAT efficiently approximates the GTR + G model; GTRMIXI uses GTRCAT during heuristic search, but the full GTR + I + G model for the final likelihood computation. Best rearrangement settings were estimated by RAxML during tree search. Bayesian analyses were performed with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001), implementing the GTR + I + G model. Three parallel runs of four incrementally heated simultaneous Markov chains were performed over 1 000 000 generations from which every 100th tree was sampled in each run. The first 200 trees were discarded, and a 90% majority rule consensus of the remaining trees was computed to obtain estimates for the probabilities that groups are monophyletic given the sequence data (posterior probabilities). The multiple sequence alignment file has been deposited in TreeBASE and is available at http://purl.org/phylo/treebase/phylows/study/TB2:S10590.
RESULTS
Phylogenetic considerations
Of the 3645 characters of the combined matrix 888 were parsimony informative (LSU: 107, ITS: 92, rpb2: 470, tef1: 219). MP analyses revealed one most parsimonious tree of length 3826 (not shown). The best ML tree (lnL = –21091.24) (Fig. 1) is similar to the MP tree except for minor differences in topology concerning Nectria berolinensis, N. lamyi and N. aurantiaca. Tree topologies of the Bayesian analyses were the same as in the ML tree. The three Bayesian runs revealed almost identical posterior probabilities. MP bootstrap support above 70%, ML bootstrap support above 70% and Bayesian posterior probabilities above 90% are illustrated (Fig. 1) at first, second and third position above or below the branches respectively. Comparison of the MP bootstrap trees from the individual genes revealed similar topologies but little backbone resolution. Differences in topology were characterized by low bootstrap support (below 65%), indicating only minor incongruence among individual gene regions. In addition MP bootstrap support was mostly higher in the combined analysis than the highest support value obtained in the single gene analyses (data not shown).
Fig. 1.
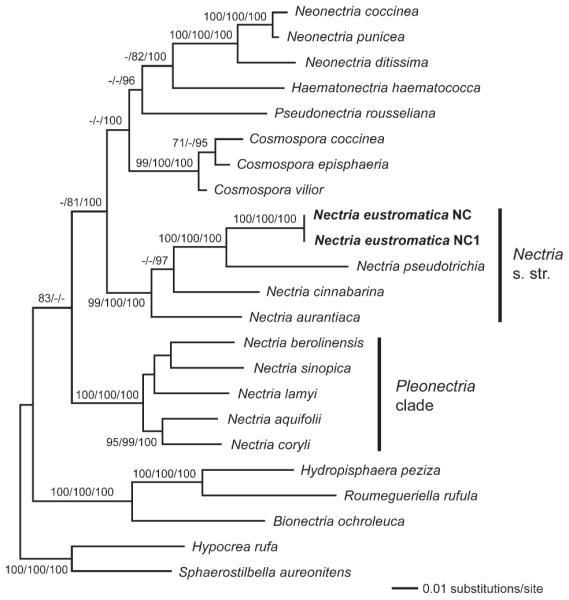
Phylogram of the best ML tree (lnL = −21091.24) revealed by RAxML from an analysis of the combined LSU-ITS-rpb2-tef1 matrix of selected Hypocreales, showing the phylogenetic position of Nectria eustromatica within Nectria sensu stricto. MP bootstrap support above 70%, ML bootstrap support above 70% and Bayesian posterior probabilities above 90% are given at first, second and third position above or below the branches.
Phylogenetic analyses place Nectria eustromatica in the Nectria sensu stricto clade with high support (Fig. 1). Sister-group relationship to N. pseudotrichia is highly supported in all analyses. Nectria is revealed as polyphyletic because it forms two distinct, highly supported clades: Nectria sensu stricto, represented in our analyses by N. cinnabarina, N. aurantiaca, N. pseudotrichia and N. eustromatica, and “Pleonectria” containing the remaining Nectria species that were included in the analyses (Fig. 1).
TAXONOMY
Nectria eustromatica Jaklitsch & Voglmayr, sp. nov. Figs. 2, 3
Fig. 2.
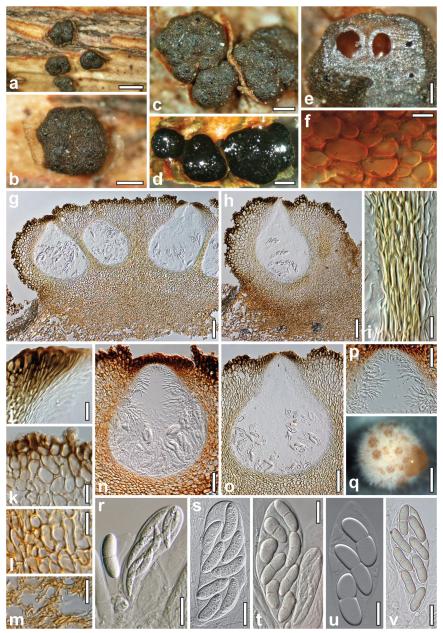
Teleomorph and sporodochia of Nectria eustromatica. a–c. Dry stromata (a. habit). d. Rehydrated stromata. e. Stroma cut horizontally showing orange-brown interior. f. Subcortical cells in cross-section in 3% KOH. g, h. Stromata in vertical section (h. uniperitheciate stroma; in lactic acid). i. Walls of two adjacent perithecia in section. j. Ostiolar apex cells. k. Cortical and subcortical tissue in section. l. Subperithecial tissue in section. m. Stroma base in section. n, o. Perithecia in section (n. in 50% glycerol; o. in lactic acid). p. Apical paraphyses. q. Sporodochium (CBS 125578, OA, 42 d). r–v. Asci and ascospores (r. immature ascus; t. mature and immature asci). Sources: a–c, e,f, r, t–v. WU 30194. d, g–p, s. WU 30195. Bars: a, q = 0.8 mm. b–d = 0.4 mm. e = 0.2 mm. f = 10 μm. g, h = 0.1 mm. i, j, l, u = 15 μm. k, r–t, v = 20 μm. m, p = 30 μm. n = 50 μm. o = 70 μm.
Fig. 3.
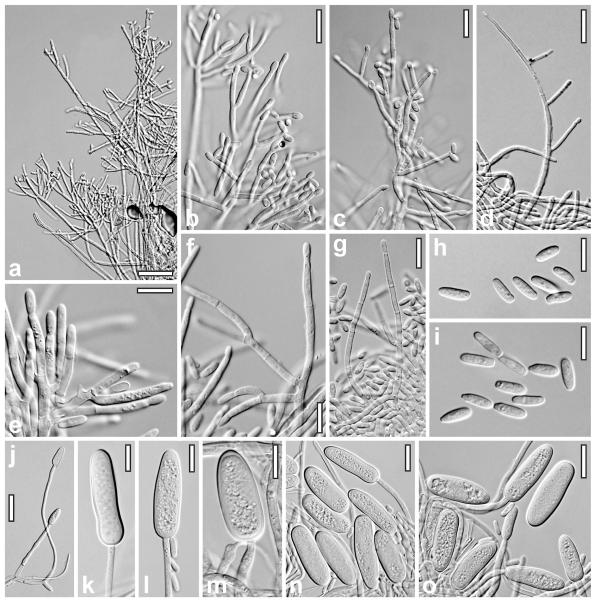
Anamorph of Nectria eustromatica. a–d. Conidiophores (a–c. MEA, 79 d; d. OA, 42 d). e–g. Phialides and conidia (e, f. PDA, 34 d; g. SNA, 26 d). h, i. Conidia (h. SNA, 26 d; i. PDA, 34 d). j. Macroconidia-forming conidiophore (OA, 42 d). k–o. Macroconidia (OA, 42 d). a–d, g, h, j–o. CBS 125578. e, f, i. CBS 121896. a–o. All at 20 ± 5 C. Bars: a = 30 μm. b, c, g, n, o = 15 μm. d, j = 20 μm. e, f, h, i, k–m = 10 μm.
MycoBank MB518506
Etymology
Eustromatica addresses the true, pseudoparenchymatous stroma.
Stromata 0.4–1.8 mm diam, pulvinata, atrofusca, pseudoparenchymatosa, 1–15 perithecia includentia. Asci (94–)100–125(−138) × (21–)24–37(−41) μm, bi- ad octospori, ellipsoidei, clavati vel saccati, unitunicati, sine apparatu apicali. Ascosporae (24–)29–37(−43) × (8–)9–12(−15) μm, biseriatae, bicellulares, oblongae vel fabiformes, hyalinae ad luteae.
Stromata (0.4–)0.7–1.4(−1.8) × (0.4–)0.5–1.1(−1.6) mm, 0.3–0.7(−0.9) mm thick (n = 50), solitary, scattered or in fascicles of up to seven, sometimes laterally fused, erumpent from bark, on a hyphal hypostroma, dark brown, orange-brown, dark gray to black, pseudoparenchymatous, with a soft consistency when fresh; pulvinate to semiglobose, nearly globose when uniperitheciate; outline variable, circular, oblong or often angular. Sides similar to the surface or lighter, dull gray to orange-brown, smooth, glabrous or slightly downy. Stroma surface convex, smooth or tubercular due to variably projecting perithecial contours, glabrous, but often appearing downy or slightly velutinous when young; when mature with 1–15 flat or convex, shiny, tarry, black ostiolar dots, (39–)57–108(−200) μm diam (n = 60), with circular, oblong or angular outline. Stroma interior lighter than the surface, orange-brown, brighter orange in 3% KOH. Stromata hydrophobic, difficult to moisten by vapor, black in water; in 3% KOH exterior unchanged, no pigment dissolved.
Stroma anatomy
Cortical layer (17–)21–45(−58) μm thick (n = 40), consisting of 1–3(−4) layers of coarse, distinct, globose or angular, dark brown to dark reddish brown cells, (9–)13–26(−34) × (7–)9–19 (−29) μm (n = 35) in section, with walls 0.5–2.5 μm thick, forming a textura angularis. Surface lacking hairs, but with erect cells or cell groups forming warts, causing the rough or velvety appearance of stromata under lower magnifications. Subcortical tissue reaching to the base of perithecia and sometimes to the stroma surface, comprising a t. angularis of roundish, angular, isodiametric to oblong cells, (7–)12–27(−39) × (6–)8–15(−24) μm (n = 35) in vertical section, (7–) 10–17(−19) × (5–)7–12(−16) μm (n = 30) in cross section, with walls 0.5–1 μm thick, yellow, orange-brown in glycerol, KOH and water; between widely spaced perithecia replaced by wide, vertically oriented hyphae. Subperithecial tissue comprising a t. epidermoidea of thin-walled, yellow cells (6–)7–19(−30) × (4–)5–12(−15) μm (n = 35), denser and darker yellow than the subcortical tissue. Basal tissue a t. intricata of thin- and thick-walled hyphae (3–)4–6(−8) μm wide (n = 35), in lower layers connecting several stromata as a subiculum, similarly pigmented or somewhat darker than subperithecial tissue. Perithecia (300–)350–460 (−490) μm high including ostioles and (235–)270–345(−380) μm wide (n = 22), sphaeroid. Peridium (17–)23–41(−50) μm thick at the base (n = 18) and (12–)21–32(−38) μm (n = 18) at the sides, yellow; at the sides of closely appressed perithecia distinct, of elongate, compressed refractive yellow cells, otherwise usually indistinctly differentiated from the surrounding pseudoparenchymatous tissue, lined inside by narrow, often collapsed, hyaline cells, 2.5–5(−8) μm wide. Ostioles (120–)125–148(−167) μm long, even with the surface or projecting 10–40(−52) μm, (24–) 26–42(−52) μm wide at the apex inside and (90–)104–144(−180) μm outside (n = 18, 18), including vertical to converging, elongate, subclavate, dark brown apical cells. Ostiole contents turning yellow in 3% KOH, filled with periphyses merging downward into acute, nearly lanceolate apical paraphyses in apical regions of the perithecia. True paraphyses absent. Asci (94–)100–125(−138) × (21–)24–37(−41) μm (n = 30), clavate, ellipsoidal or saccate, with a minute stipe or acute base, without a differentiated apical structure; entirely filled with (2–)4–8 ascospores in biseriate arrangement. Ascospores (24–)29–37(−43) × (8–)9–12 (−15) μm, l/w (1.7–)2.6–3.6(−4.5) (n = 60), hyaline to yellowish, first unicellular, falcate or sigmoid with acute ends when immature, becoming bicellular, straight or slightly curved, allantoid to bean-shaped, with broadly rounded ends, thick-walled (ca. 1 μm); septum central, not constricted; perispore thin, hyaline, delicately verruculose in 3% KOH when old.
Cultures and anamorph
Nectria eustromatica grows on all agar media tested, fastest on PDA and OA. It sporulates on all media except CMD. Macroconidia were found only on sporodochia on MEA and OA.
On SNA at 20 ± 2 C colony colorless, thin, circular, margin ill defined. No pigment, no distinct odor formed. After 2 wk conidia produced in minute wet heads; later in white granules consisting of compact, dense aggregates of conidiophores. Conidia only rarely > 10 μm long.
On OA at 20 ± 2 C colony whitish, pale rosy to pale brown with gray margin; odor unpleasant, acidic; after 4–5 wk plate covered by mycelium. In 1–6 wk numerous white fluffy tufts appearing in the colony center, spreading across the colony. Tufts partly turning into sporodochia within several weeks; conidiophores becoming fertile after 5–6 wk.
On MEA after 4 mo at 15 C colony colorless, without a distinct odor; conidia produced in mucous, carrot-colored drops on sporodochia; macroconidia produced within 1 y. At 20–25 C colony white to yellowish, with short spiny aerial hyphae; white tufts appearing after approximately 1 mo. After 2.5–3 mo minute white tufts and pale brownish sporodochia up to ca. 4 mm diam present.
On PDA at 20–25 C growth fast, after 4–5 wk plate covered by mycelium. Colony whitish to dull yellow, lobed. Conidiation after 5 wk in mostly roundish, white shrubs, fluffy tufts, spots or on sporodochia.
Sporodochia 0.3–3.5(−4) mm diam, compact, pulvinate, semiglobose, ellipsoidal to subglobose, white, pale reddish brown, to carrot-colored (on MEA), pseudoparenchymatous, with a loose white tomentum of aerial hyphae and conidiophores on surfaces. Conidiophores either solitary on surface hyphae, in minute white tufts or densely aggregated on the surface of sporodochia; erect, more or less fan- or broom-shaped, consisting of a straight main axis mostly 3–5 μm wide, attenuated upward to 2–3.5 μm terminally, cylindrical or with widening to 6–8 μm in age, smooth, with age becoming thick-walled with outer wall swelling in KOH; rarely with small rounded warts. Main axis unbranched or with monochasial, 1- to few-celled branches 3–4.5 μm wide, at several levels, loosely and asymmetrically arranged at acute angles, rarely perpendicular; branches rarely paired or verticillate. Phialides terminal on branches of similar width, solitary or in groups of two, divergent, rarely parallel, (7–)10–20(−37) × (2.0–)2.2–2.7(−3.0) μm, l/w (3.0–)4.2–8.0(−12.8) (n = 60; from PDA and SNA), cylindrical, straight or slightly curved, sometimes slightly constricted at the base. Conidia numerous, amassing in colorless, brownish to carrot-colored, turbid drops up to 0.4 mm diam, (5.5–)6.8–9.5(−13.8) × (2.0–)2.5–2.8(−3.5) μm, l/w (2.0–)2.6–3.7(−5.1) (n = 78; from MEA, PDA and SNA; with highest variability in size on SNA); hyaline, unicellular, oblong to cylindrical, straight or slightly curved, smooth, eguttulate or with inconspicuous minute guttules, often with a distinct, truncate abscission scar. Non-phialidic macroconidia produced on the same conidiophores in basal regions or terminally on long narrow hyphae 2–3.5 μm wide; macroconidia formed solitarily, (20–)27–38(−54) × (8.7–)9.7–12.5 (−14.8) μm, l/w (1.5–)2.4–3.5(−4.1) (n = 55, from MEA and OA), hyaline, oblong, cylindrical or narrowly ellipsoidal, straight or curved, smooth, with walls 0.8–1.7 μm thick, eguttulate, without a scar.
Habitat
on recently dead standing branches/trunks of Hippocrepis (Coronilla) emerus.
Distribution
Southern Europe, collected in Croatia and Italy.
Holotype
CROATIA, PRIMORSKO-GORANSKA, Opatija, Mošcenička Draga, village area, on dead twigs of Hippocrepis emerus, soc. Cucurbitaria coronillae and some immersed pyrenomycetes, 29 Mar 2007, W. Jaklitsch & H. Voglmayr, W.J. 3079 (WU 30194; culture NC = CBS 121896).
Additional material examined
ITALY, LAZIO, Bagnaia, Prov. Viterbo, at Villa Lante, on Hippocrepis emerus, 28 Jul 2009, W. Jaklitsch, H. Voglmayr & W. Gams (WU 30195; culture NC1 = CBS 125578).
DISCUSSION
Nectria eustromatica is a member of Nectria sensu stricto (Fig. 1). For a long time the genus Nectria was conceived as fungi forming bright-colored, superficial perithecial ascomata with one-septate hyaline ascospores in unitunicate asci and devoid of true paraphyses. Scolecosporous taxa were classified in Scoleconectria Seaver (see Booth 1959) and Ophionectria Sacc. (Rossman 1977), those with muriform ascospores in Pleonectria Sacc. or Thyronectria Sacc. (Seeler 1940, Booth 1959). Later ascomatal wall structure and anamorphs were given superior significance in the definition of genera by Samuels and Rossman (1979). Rossman (1983) added phragmosporous taxa and later (Rossman 1989) dictyosporous taxa to the genus Nectria when she defined the Nectria cinnabarina group. The species of this group are characterized by large, often warted, often collabent, more or less red, KOH+ ascomata with a two-layered wall, sometimes covered by yellow or greenish scurf, aggregated in cespitose clusters on an often well developed, pseudoparenchymatous hypostroma, and by anamorphs placed in the form genera Tubercularia Tode, Stilbella Lindau, Gyrostroma Naumov or Zythiostroma Höhn. ex Falck. This group was refined by Rossman et al. (1999) and was known as Nectria sensu stricto until Hirooka et al. (2009) determined that Nectria is paraphyletic and falls into two major clades; thus Nectria sensu stricto becomes restricted to species with Tubercularia anamorphs centering around the type species of Nectria, N. cinnabarina. Species with the pycnidial Gyrostroma and Zythiostroma anamorphs are included in the second principal clade, here called “Pleonectria”.
Nectria eustromatica is a member of Nectria sensu stricto according to the most recent circumscription. All other species of Nectria sensu stricto differ from N. eustromatica by discrete perithecia. Perithecia of Nectria cinnabarina sometimes may be laterally fused, as shown by Seifert (1985, p 100), and have thick-walled clavate ostiolar cells similar to those of N. eustromatica, as shown by the same author. Our fungus may be interpreted as a result of a development of the thick pseudoparenchymatous wall of N. cinnabarina to a compact stroma, as an upward extension of the pseudoparenchymatous hypostroma that merged with the outer parts of the perithecial wall. Nectria pseudotrichia, which, according to the phylogenetic analysis, is a sister species of N. eustromatica, differs by muriform ascospores and the stipitate, synnematous anamorph Tubercularia lateritia (Berk.) Seifert from N. eustromatica. Also Nectria aurantiaca differs by a similar synnematous anamorph. No anamorph of N. eustromatica has been seen in nature. The anamorph of N. eustromatica formed in culture is assignable to either Tubercularia or Stilbella devoid of synnemata. Conidiophores of N. eustromatica with repeatedly monochasial branching are similar to those of the Nectria aurantiaca and N. pseudotrichia anamorphs (Seifert 1985, p 105, p 123, FIG. 39d; Booth 1959, p 30), whereas conidiophores of Tubercularia vulgaris, the anamorph of N. cinnabarina, are acropleurogenous in contrast to those of our fungus, which has branches of almost equal length as the elements of the axis. The formation of macroconidia has not been reported for any species currently known to belong to Nectria sensu stricto. They are produced later than the phialoconidia, often only after several months, while the latter are formed during a long period, usually still at times when macroconidia appear. The macroconidia, albeit being distinctly larger, resemble phialoconidia of N. aurantiaca (Booth 1959). Interestingly, catalinensis C.E. Lima, described from Gleditsia in Argentina (Lima et al. 1988), forms similar but smaller macroconidia (21–24.5 × 11.5–15 μm) in culture. Nectria catalinensis, N. balansae Speg. and N. sordida Speg. are also similar to N. eustromatica in forming pseudoparenchymatous stromata and ascospores of similar size. However stromata of these species, all described from Argentina, are red or reddish-brown and have a tubercular surface due to partly projecting perithecia. Nectria catalinensis and N. balansae differ from N. eustromatica also in striate ascospores, and N. sordida in distinctly smaller perithecia (Lima et al. 1988, Samuels and Brayford 1994). The phylogenetic placement of N. catalinensis and N. sordida is unknown, while N. balansae is shown to be only distantly related to N. eustromatica phylogenetically, based on a LSU sequence. The ascospores of N. eustromatica, although wider, are similar to those of the Indian Peethambara sundara Subram. & Bhat in length and shape (Rossman et al. 1999). However the latter fungus has free, yellow perithecia and belongs to the Bionectriaceae. Its anamorph produces large bicellular conidia on synnemata and belongs to genus Didymostilbe Henn. (Seifert 1985).
Stromata of Nectria eustromatica occur specifically on stems of Hippocrepis emerus, mostly on basal parts, maturing from there. Immature stromata of Cucurbitaria coronillae (Fr.) Sacc., which usually occur in large numbers on the same branches, superficially resemble those of N. eustromatica. However a cross section of a N. eustromatica stroma exposing the orange or orange-brown interior reveals its true nature. The superficial similarity of these two fungi is probably the reason why N. eustromatica apparently has escaped notice. The overall appearance is that of a dothidealean fungus. No hint toward a description of this fungus in such a genus has been found, except for Dothidea coluteae Berk. & M.A. Curtis, described from twigs of a Colutea sp. in Pennsylvania, USA. The holotype (K 164150) of this fungus was examined and found to be a typical Dothidea, similar to D. sambuci, with bicellular, yellowish brown ascospores, 17–23 × 6.5–9.5 μm, of slightly unequal cells, in bitunicate asci.
ACKNOWLEDGMENTS
We thank Walter Gams for hospitality and excursion support in Italy; the fungarium curators of K, B. Spooner and B. Aguirre-Hudson for loan of type material, CBS for cultures of Nectria aurantiaca and N. pseudotrichia, EU SYNTHESYS for a grant (GB-TAF-4625) to HV for a research stay at Royal Botanic Gardens Kew enabling him to collect Nectria aquifolii and the Austrian Research Fund (FWF; projects P19143, P22081) for financial support.
LITERATURE CITED
- Booth C. Studies of Pyrenomycetes IV. Nectria I. Mycol Pap. 1959;73:1–115. [Google Scholar]
- Chaverri P, Samuels GJ. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud Mycol. 2004;48:1–116. 2003. [Google Scholar]
- de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous Basidio-mycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. doi:10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. doi:10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka Y, Rossman AY, Chaverri P. Systematics of the genus Nectria based on a six-gene phylogeny [MSA meeting 2009 abstract] Inoculum. 2009;60:22. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM. European species of Hypocrea I. The green-spored species. Stud Mycol. 2009;63:1–91. doi: 10.3114/sim.2009.63.01. doi:10.3114/sim.2009.63.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.3852/mycologia.97.6.1365. doi:10.3852/mycologia.97.6.1365. [DOI] [PubMed] [Google Scholar]
- Lima CE, Forchiassin F, Ranalli ME. Systematic and biological study of Hypocreales of Argentina IV. Nectria catalinensis sp. nov. Nova Hedwig. 1988;46:149–156. [Google Scholar]
- Liu YL, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Riethmüller A, Voglmayr H, Göker M, Weiß M, Oberwinkler F. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia. 2002;94:834–849. doi: 10.1080/15572536.2003.11833177. doi:10.2307/3761698. [DOI] [PubMed] [Google Scholar]
- Rossman AY. The genus Ophionectria (Euascomycetes, Hypocreales) Mycologia. 1977;69:355–391. doi:10.2307/3758661. [Google Scholar]
- Rossman AY. The phragmosporous species of Nectria and related genera. Mycol Pap. 1983;150:1–164. [Google Scholar]
- Rossman AY. A synopsis of the Nectria cinnabarina-group. Mem New York Bot Garden. 1989;49:253–265. [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes) Stud Mycol. 1999;42:1–248. [Google Scholar]
- Samuels GJ, Brayford D. Species of Nectria (sensu lato) with red perithecia and striate ascospores. Sydowia. 1994;46:75–161. [Google Scholar]
- Samuels GJ, Petrini O, Kuhls K, Lieckfeldt E, Kubicek CP. The Hypocrea schweinitzii complex and Trichoderma sect. Longibrachiatum. Stud Mycol. 1998;41:1–54. [Google Scholar]
- Samuels GJ, Rossman AY. Conidia and classification of the nectrioid fungi. In: Kendrick WB, editor. The Whole Fungus. Vol. 1. 1979. pp. 167–179. [Google Scholar]
- Seeler EV. A monographic study of the genus Thyronectria. J Arnold Arboretum Harvard Univ. 1940;21:429–460. [Google Scholar]
- Seifert KA. A monograph of Stilbella and some allied Hyphomycetes. Stud Mycol. 1985;27:1–235. [Google Scholar]
- Stamatakis E. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi:10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Prosthecium species with Stegonsporium anamorphs on Acer. Mycol Res. 2008;112:885–905. doi: 10.1016/j.mycres.2008.01.020. doi:10.1016/j.mycres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Werle E, Schneider C, Renner M, Völker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. doi:10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press; Dan Diego: 1990. pp. 315–322. [Google Scholar]