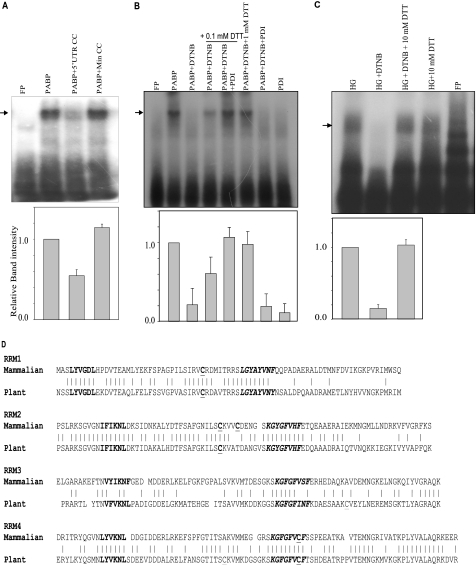
FIGURE 6.
PABP bind to the insulin 5′ UTR. A, competitive RNA-EMSA was performed as described under “Experimental Procedures.” The sample in each lane is as indicated. The radiolabeled insulin 5′ UTR (FP) was incubated with bacterially expressed recombinant PABP (100 ng) in the presence of the unlabeled insulin 5′ UTR or mutant 5′ UTR RNA as competitor. The gel-shifted bands were quantitated densitometrically, and expressed with respect to only PABP without competitor set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. B, RNA-EMSA was performed to assess the role of free cysteines in the RNA binding activity. The radiolabeled insulin con1 probe was incubated with PABP, PABP oxidized with DTNB, and the oxidized PABP reduced with varying amounts of DTT or DTT and PDI (200 ng). The gel-shifted bands were quantitated densitometrically, and expressed with respect to only PABP without competitor set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. C, the radiolabeled con1 insulin 5′ UTR were incubated with islet extracts, extracts oxidized with DTNB, and oxidized extracts reduced with DTT. The gel-shifted bands of HG, HG-DTNB, and HG + DTNB + DTT were quantitated densitometrically, and expressed with respect to the HG extract set to 1 (lower panel). The graph represents the average of four independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. D, amino acid sequence comparison of the RRMs of PABP from mammals and plants. The conserved hexamer (bold) and octamer (bold italics) in RRMs, as well as the cysteine residues (bold underlines) are indicated.