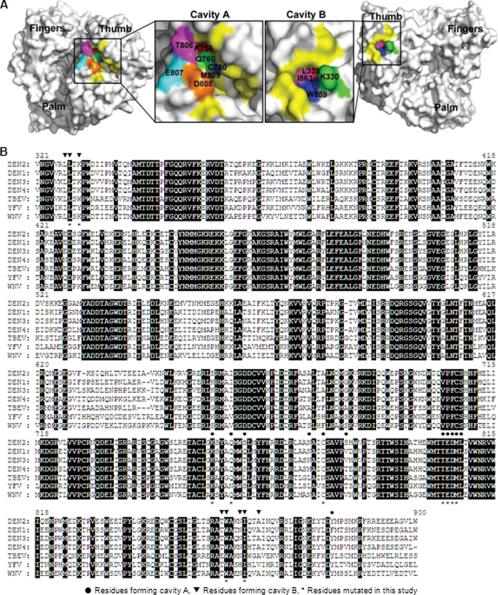
FIGURE 1.
Structure and sequence analyses of cavities A and B from DENV RdRp. A, cavities A and B in DENV-3 RdRp structure. The crystal structure of DENV-3 RdRp (Protein Data Bank entry 2J7U) was used to illustrate cavity A (left panel) and cavity B (right panel). Amino acids constituting the cavities are labeled in yellow or other colors. Residues selected for mutagenesis analysis are colored as follows: for cavity A, Lys-756 (red), Gln-760 (green), Cys-780 (blue), Thr-806 (magenta), Glu-807 (cyan), Asp-808 (orange), and Met-809 (tint); for cavity B, Leu-328 (red), Lys-330 (green), Trp-859 (blue), and Ile-863 (pink). The images of RdRp were produced using PyMOL. B, amino acid sequence alignment of partial RdRp region from the four serotypes of DENV and other flaviviruses. The RdRp amino acid sequences of DENV-1, DENV-2, DENV-3, DENV-4, WNV, yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV) are derived from GenBankTM accession numbers U88535, AY037116, M93130, AY947539, AF404756, X03700, and AF069066, respectively. Identical amino acids among all RdRps are shaded. The numbering of amino acid sequence is based on DENV-2. The residues involved in the formation of cavity A and B are indicated by ● and ▾ above the sequence, respectively. The mutated residues in this study are indicated by * below the sequence.