FIGURE 4.
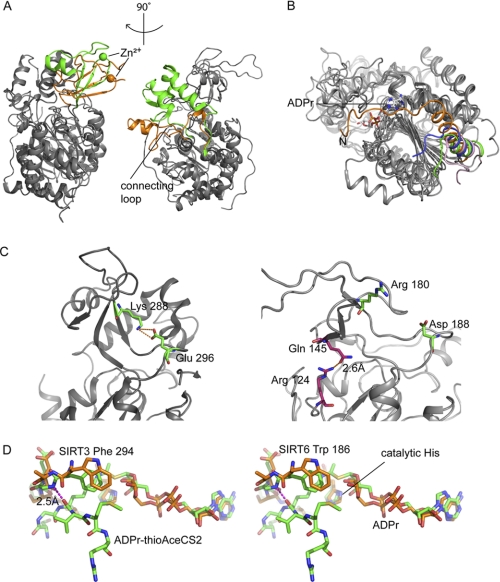
SIRT6·ADPr structure compared with other solved human SIRTs. A, SIRT6 structure (PDB code 3PKI, orange) superimposed onto SIRT3 (PDB code 3GLR, green). The left panel highlights differences in the zinc-binding domain, and the right panel shows a missing helix bundle. B, comparison of the N-terminal loop of SIRT6 (3PKI, orange), SIRT2 (1J8F, pink), SIRT3 (3GLR, green), and SIRT5 (2B4Y, blue); SIRT6 contains a long loop covering the NAD+ binding site. C, SIRT6 lacks the conserved salt bridge. Left, SIRT3 (3GLR) structure with conventional salt bridge in the sirtuin family. Right, the green pair (Arg180 and Asp188) is where the bridge would be predicted to form on the SIRT6 structure according to other sirtuin crystal structures. Pink pair (Arg124 and Gln145), the actual hydrogen bonding pair found in SIRT6 structure. D, stereo diagram of flipped FGEXL (WEDsL) loop of SIRT6 (PDB code 3PKI, orange) complexed with ADPr (orange) compared with SIRT3 (PDB code 3GLT, green) with an ADPr·thioAceCS2 (green) complex.