Figure 6.
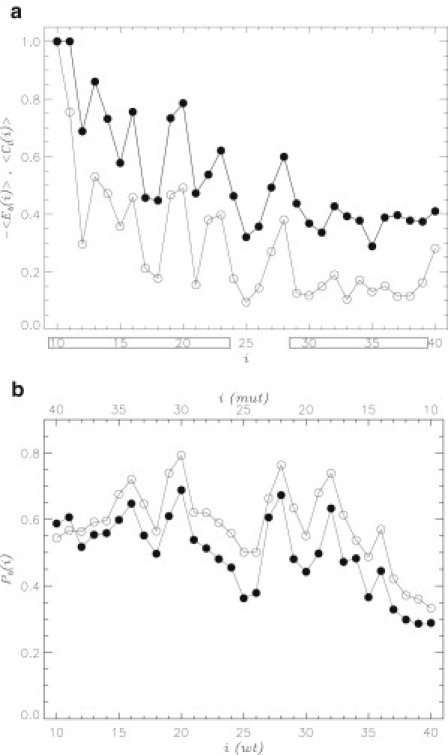
(a) Number of contacts formed by amino acid i in Aβ dimer with naproxen ligands (solid circles). Energy of interactions between amino acid i and ligands are represented by open circles. Both quantities are computed using REMD at 330 K and normalized. The Nt and Ct terminals are boxed. (b) Probability of naproxen binding to amino acid i in Aβ monomer computed using AutoDock: WT Aβ (solid circles), reversed Aβ mutant (open circles, Fig. 1b). Amino acid numbers for the mutant sequence are shown on top in a backward direction (from 40 to 10). Both panels implicate ligand-amino acid interactions as a main factor controlling the location of ligand binding sites along Aβ sequence.
