This study shows that the Per-ARNT-Sim-like MEKHLA domain of the Arabidopsis class III homeodomain leucine zipper transcription factor REVOLUTA acts as a negative regulator of this protein by inhibiting its dimerization.
Abstract
Class III homeodomain leucine zipper (HD-ZIP III) transcription factors regulate critical developmental programs in plants; these include leaf polarity, polarity along the shoot-root axis, and stem cell specification and proliferation. One of the defining features of HD-ZIP III proteins is the presence of a Per-ARNT-Sim-like (PAS-like) MEKHLA domain at the C terminus. PAS-like domains are known to respond to a variety of chemical and physical stimuli. Here, we provide evidence that the MEKHLA domain acts as a negative regulator of Arabidopsis thaliana HD-ZIP III REVOLUTA activity. Based on experiments in yeast and plants, we propose a model in which the MEKHLA domain inhibits dimerization through a sequence-independent steric masking mechanism. This inhibition is relieved in response to a cellular signal that requires the C terminus of the MEKHLA domain for its perception. Overexpression experiments suggest that this signal is unequally distributed and/or sensed in the plant. Our data show that the function of the REVOLUTA MEKHLA domain differs among other HD-ZIP III family members; this difference may explain the genetic differences that have been observed among family members. This finding, combined with our phylogenetic analysis, suggests that REVOLUTA is the latest type of HD-ZIP III protein to have evolved in land plants.
INTRODUCTION
Class III homeodomain leucine zipper proteins (HD-ZIP III proteins) are plant-specific transcription factors that play prominent roles in plant development. In Arabidopsis thaliana, their activity establishes the shoot pole of the shoot-root axis in embryonic development (Grigg et al., 2009; Smith and Long, 2010), establishes the adaxial (upper) domain of the leaf primordium (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005; Ochando et al., 2006, 2008), promotes meristem formation (Talbert et al., 1995; McConnell and Barton, 1998; Otsuga et al., 2001; Hawker and Bowman, 2004; Prigge et al., 2005), regulates growth within the shoot apical meristem (Green et al., 2005; Williams et al., 2005; Ochando et al., 2006, 2008; Zhou et al., 2007), and patterns the vasculature (Baima et al., 2001; Zhong and Ye, 1999; Carlsbecker et al., 2010). The HD-ZIP III proteins are conserved among land plants (Sakakibara et al., 2001; Floyd et al., 2006; Prigge and Clark, 2006) and have been shown to play similar developmental roles in other flowering plants (Juarez et al., 2004; McHale and Koning, 2004).
The HD-ZIP proteins are named for the combination of homeodomain and leucine zipper domains at their N terminus (Ruberti et al., 1991; Schena and Davis, 1992). Unlike animal homeodomain proteins, HD-ZIP proteins bind DNA as dimers in vitro, where they bind a palindromic sequence (Sessa et al., 1993, 1997, 1998). The loop between helices one and two is critical in determining whether the homeodomain binds as a monomer or dimer; substitution of the loop between helices one and two with the corresponding loop from the ENGRAILED homeodomain allows HD-ZIP proteins to bind DNA as monomers (Tron et al., 2004).
Dimerization of HD-ZIP III proteins in vivo is regulated by a family of small proteins that consist almost entirely of leucine zipper sequence (Wenkel et al., 2007; Kim et al., 2008). These LITTLE ZIPPER (ZPR) proteins interact with HD-ZIP III proteins in vitro and prevent DNA binding. Overexpression of ZPR proteins causes phenotypes similar to those seen in loss-of-function HD-ZIP III mutants. These observations are the basis for a model in which ZPR proteins act to negatively regulate HD-ZIP III activity by preventing their dimerization.
The structure of HD-ZIP III proteins suggests additional mechanisms of posttranslational regulation. In addition to the homeodomain and leucine zipper domain, HD-ZIP III proteins contain, in order from N terminus to the C terminus, a steroidogenic acute regulatory protein lipid transfer domain (START), a homeodomain-START associated domain (HD-SAD), and a MEKHLA domain (Ponting and Aravind, 1999; Mukherjee and Bürglin, 2006). START domains in animals bind cholesterol, phospholipids, and carotenoids (Radauer et al., 2008). In plants, the PYR/PYL group of START domains binds ABA and appears to act as an ABA receptor (Ma et al., 2009; Park et al., 2009). It is not known what ligand, if any, binds to the HD-ZIP III START domain.
The MEKHLA domain is a member of the Per-ARNT-Sim (PAS) domain superfamily. PAS domains are signal sensors that regulate a wide range of signal transduction pathways in all kingdoms of life. They respond to a variety of chemical and physical stimuli and regulate the activity of covalently linked effector domains, such as kinases, cyclases, ion channels, and transcription factors. Möglich et al. (2009) propose that the typical role of PAS domains is to modulate the mono/dimeric state of effector domains through conformational changes triggered by a signal. The canonical PAS fold consists of a central five-stranded β-sheet flanked by several α-helices with the β-sheet playing a central role in cofactor binding and signal propagation.
The HD-ZIP III proteins are unique in the Arabidopsis proteome in carrying a MEKHLA domain. The closely related class IV HD-ZIP proteins have a similar domain structure to that of the class III proteins except that they lack a MEKHLA domain. Proteins that consist solely of a single MEKHLA domain exist in the blue-green algae (prokaryotes) and in the green alga Chlamydomonas reinhardtii (Mukherjee and Bürglin, 2006).
Of the five HD-ZIP III genes, (REVOLUTA [REV], PHABULOSA [PHB], PHAVOLUTA [PHV], INCURVATA4/CORONA [ICU4/CNA], and ATHB8), only rev single mutants have a readily observable mutant phenotype (Talbert et al., 1995; Zhong and Ye, 1999; Ratcliffe et al., 2000, Otsuga et al., 2001). Loss of REV function leads to failure to produce axillary meristems and functional floral meristems. It also leads to alteration of vascular patterning within the stem. When rev mutations are combined with loss-of-function mutations in the PHB and PHV genes, the resulting triple mutant embryos show abnormal pattern formation (Emery et al., 2003; Prigge et al., 2005). They frequently form a single radially symmetric cotyledon instead of the normal pair of ad/abaxially polarized cotyledons. Defects in polar development of leaves and floral organs is also seen in plants with combinations of rev, phb, and phv mutations; abaxial (lower) fates replace adaxial (upper) fates in these higher-order mutants. Thus, PHB and PHV provide redundant activities for some but not all REV activity.
ICU4/CNA and ATHB8 are less closely related to REV than PHB and PHV are. Nevertheless, ICU4/CNA (but not ATHB8) acts redundantly with REV in establishing normal embryo patterning (Green et al., 2005; Prigge et al., 2005). ICU4/CNA also acts redundantly with PHB and PHV (but not REV) in regulating meristem size. Surprisingly, ICU4/CNA and ATHB8 act oppositely to REV in regulating the formation of lateral shoot and floral meristems. Thus, HD-ZIP III proteins display a complex pattern of interactions that include both opposing and redundant interactions.
Much of the spatial regulation of HD-ZIP III expression is established through the action of the mir165/166 family of microRNAs (miRNAs; Rhoades et al., 2002; Mallory et al., 2004; Williams et al., 2005; Zhou et al., 2007; Carlsbecker et al., 2010). The mir165/166 miRNAs trigger the degradation of HD-ZIP III mRNAs, thereby limiting HD-ZIP III mRNA accumulation to a subset of cells in which they are transcribed. Gain-of-function mutations in four of five HD-ZIP III genes (REV, PHB, PHV, and ICU4/CNA, but not ATHB8) have been isolated (McConnell and Barton, 1998, 2001; Ochando et al., 2006, 2008). These mutations are located in the miRNA complementary site of the transcript and render the mRNA miRNA resistant, causing ectopic expression of the corresponding mRNAs. These mutants exhibit dramatic alterations in organ polarity, embryonic patterning, and vascular patterning.
Despite being subject to miRNA regulation by the same subset of miRNAs, the five Arabidopsis HD-ZIP III genes are expressed in different, overlapping patterns (Baima et al., 1995; McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005; Williams et al., 2005). Hence, the expression domain of each of the five HD-ZIP III genes is determined by their promoters as well as by localized miRNA action.
In this study, we identify and characterize the role of the MEKHLA domain in modulating REV dimerization. Our data suggest a mechanism in which the N-terminal region of the MEKHLA domain is sufficient to cause conformational changes that inhibit dimer formation. The inhibition is relieved in response to a signal that requires the C terminus of the MEKHLA domain for its perception. Overexpression analyses in planta indicate that such a signal is not equally distributed or sensed in all cell types. This biological switch lies upstream of previously known mechanisms of REV posttranslational regulation.
RESULTS
The MEKHLA domain is a conserved domain located at the C terminus of all HD-ZIP III proteins (Mukherjee and Bürglin, 2006). The MEKHLA domain consists of a PAS-like domain flanked by a conserved N-terminal domain (Mukherjee and Bürglin, 2006). The PAS-like domain is predicted to have a similar secondary structure to that of well-defined PAS domains, such as that of Sinorhizobium meliloti and Azotobacter vinelandii, whose structures are known (Figures 1A and 1B) (Möglich et al., 2009). The N-terminal flanking region has weak similarity to the WD-40 domain (see Supplemental Figure 1 online); therefore, we named it MEKHLA WD40-like (M-WD40) (note, however, that it is not at all clear that this region will have a similar structure to WD-40 domains, which consist of several strands of β-sheet rather than α-helix). The PAS domain in turn contains two regions of sequence conservation that have been named the S1 and S2 domains (Zhulin et al., 1997).
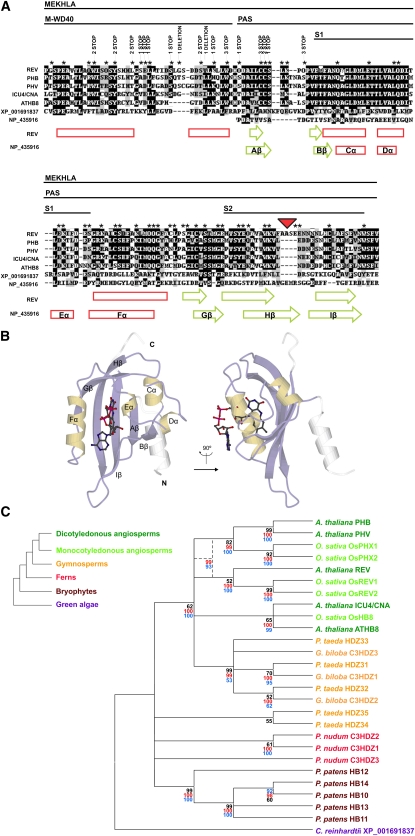
Figure 1.
The REV MEKHLA Domain Belongs to the PAS Domain Superfamily.
(A) Multiple sequence alignment of the Arabidopsis (REV, PHB, PHV, CNA, and ATHB8) and C. reinhardtii (XP_001691837) MEKHLA domains and the PAS domain of the S. meliloti NP_435916 protein. Conserved subdomains are indicated by lines on top of the alignment. A schematic representation of the secondary structure as determined in the NP_435916 protein structure and predicted in REV is drawn below the alignment. Green arrows indicate β-sheets, and red rectangles indicate α-helices. Asterisks indicate REV residues whose codons can be mutated into a stop codon by a single point mutation. The results of the REV MEKHLA domain mutagenesis performed for the reverse Y2H assay are annotated over the asterisk line. The red triangle indicates a four–amino acid stretch that is REV unique.
(B) Crystal structure of the PAS A domain of the A. vinelandii NifL protein. This PAS domain binds a flavin adenine dinucleotide cofactor. It is unknown if the REV MEKHLA domain binds any cofactor. Drawing reproduced with permission from Möglich et al. (2009).
(C) Neighbor-joining bootstrap consensus tree generated from the multiple sequence alignment of the Arabidopsis, Oryza sativa, Ginkgo biloba, Pinus taeda, Psilotum nudum, Physcomitrella patens, and C. reinhardtii MEKHLA domains. The tree was condensed with a cutoff value of 50%. Black numbers indicate bootstrap values. A neighbor-joining and a maximum parsimony bootstrap consensus tree were also constructed using full-length HD-ZIP III sequences if available. Red and blue numbers indicate bootstrap values higher than 50% for the neighbor-joining and maximum parsimony tree, respectively, in nodes that are conserved between the MEKHLA domain and the full-length HD-ZIP III trees. Dashed lines indicate a phylogenetic node resolved only by the full-length HD-ZIP III trees. The phylogram on the left illustrates the relationships between land plant clades.
The MEKHLA domain is predicted to adopt a similar structure to PAS domains (Mukherjee and Bürglin, 2006). PAS domains are sensors able to perceive chemical and physical stimuli (Möglich et al., 2009). After sensing a stimulus, a signal originates within the PAS domain and propagates to an effector domain through a short C- or α-helical linker. Similarly, the MEKHLA domain might regulate the activity of HD-ZIP III proteins by responding to a biological stimulus. The PAS region of the MEKHLA domain might detect such a stimulus and transmit a signal to another domain of the HD-ZIP III protein through the M-WD40 subdomain.
The MEKHLA Domain Is Not Required for REV Activity
To determine if the MEKHLA domain is required for REV function, we compared the ability of full-length REV to complement a rev-1 mutant with that of REV lacking the MEKHLA domain (REV-ΔMEKHLA). The most striking morphological phenotype of homozygous rev-1 mutants is their lack of cauline paraclades (branches born in the axils of leaves on the inflorescence stem) and their frequent failure to form pistils (Talbert et al., 1995). REV and REV-ΔMEKHLA proteins, constitutively expressed under the cauliflower mosaic virus 35S promoter (35S), are nearly indistinguishable in their ability to restore cauline branch and pistil production to rev-1 plants (Figure 2A). In addition, the leaves of 35S-REV-ΔMEKHLA; rev-1 plants are severely curled up (Figure 2C). This curling was not observed in 35S-REV; rev-1 plants (Figure 2B). Upward leaf curling is a hallmark of ectopic HD-ZIP III expression and is commonly seen when HD-ZIP III genes are resistant to regulation by miRNAs (Juarez et al., 2004; Mallory et al., 2004; Ochando et al., 2006, 2008). Thus, REV-ΔMEKHLA causes upward leaf curling even though this construct is under normal miRNA control.
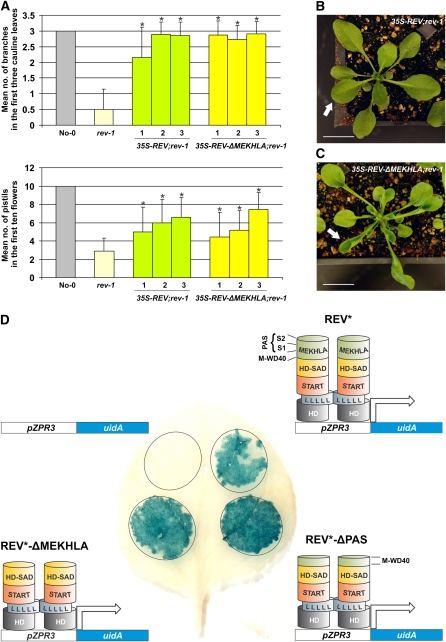
Figure 2.
The REV MEKHLA Domain Is Not Necessary for REV Activity in Vivo.
(A) Arabidopsis rev-1 plants were transformed with 35S-REV or 35S-REV-ΔMEKHLA constructs. Three independent T2 lines (1 to 3) for each genotype were examined for the number of branches in the first three cauline leaves and for the number of pistils in the first 10 flowers. rev-1 and wild-type plants (No-0) were also analyzed as controls. Standard deviations (error bars) were calculated from more than 30 individuals. Asterisks indicate transgenic lines that are statistically different from rev-1 (Student's t test, P < 0.05). A combined analysis of the lines 1 to 3 of 35S-REV; rev-1 and 35S-REV-ΔMEKHLA; rev-1 plants shows no statistically significant difference (Student’s t test, P > 0.05) in the number of pistils and a mild difference (Student’s t test, P = 0.026) in the number of branches. Lines 2 and 3 have no statistically significant difference (Student's t test, P > 0.05) in the number of branches.
(B) A representative individual (6 weeks old) of 35S-REV; rev-1 transgenic plants. The white arrow points to a normally flattened leaf. Bar = 100 mm.
(C) A representative individual (6 weeks old) of 35S-REV-ΔMEKHLA; rev-1 transgenic plants. The white arrow points to an abnormally curled up leaf. Bar = 100 mm.
(D) β-Glucuronidase expression in leaf abaxial (lower) epidermal cells of tobacco transiently transformed with pZPR3-uidA and 35S (empty vector), pZPR3-uidA, and 35S-REV*, pZPR3-uidA, and 35S-REV*-ΔPAS, or pZPR3-uidA and 35S-REV*-ΔMEKHLA constructs. miRNA-resistant REV is designated REV*. REV homodimerization through the leucine zipper domain is schematically represented by two strings of Leu residues (L) touching each other.
To further test the ability of REV-ΔMEKHLA to function, we tested whether deleting parts of the MEKHLA domain would affect REV ability to act as a transcriptional activator. We did this by overexpressing full-length REV, REV-ΔMEKHLA, or REV-ΔPAS in the presence of a construct carrying the ZPR3 gene promoter (pZPR3) fused to the reporter gene uidA (uidA encodes the enzyme β-glucuronidase) (Wenkel et al., 2007). REV binds to an intron within pZPR3 and transcriptionally activates ZPR3 (M.K. Barton and S. Wenkel, unpublished data). We performed our analyses in tobacco (Nicotiana tabacum) leaf abaxial (lower) epidermal cells, where neither REV nor ZPR3 is expected to be expressed (McHale and Koning, 2004; Wenkel et al., 2007). In this experiment, REV mRNAs were made miRNA resistant through a silent mutation in the miRNA binding site to bypass the miRNA machinery active in this cell type (constructs carrying mutations that render HD-ZIP III genes miRNA resistant are designated HD-ZIP III*). All three REV* protein forms activated uidA (Figure 2D), demonstrating that the MEKHLA domain is not necessary for REV to act as a transcriptional activator. Overall, these data suggest that the REV MEKHLA domain is not required for REV activity per se but instead plays a regulatory role.
The MEKHLA Domain Inhibits REV Dimerization in Yeast
It has been speculated that a general role for PAS domains is to modulate protein hetero- or homodimerization (Möglich et al., 2009). To determine if the MEKHLA domain might play a role in regulating dimerization, we performed yeast two-hybrid (Y2H) analyses on full-length REV and REV lacking the MEKHLA domain (REV-ΔMEKHLA). Interestingly, full-length REV did not homodimerize in Y2H assays. By contrast, REV-ΔMEKHLA strongly homodimerized in Y2H experiments (Figure 3A). Thus, the MEKHLA domain prevents REV homodimerization in yeast.
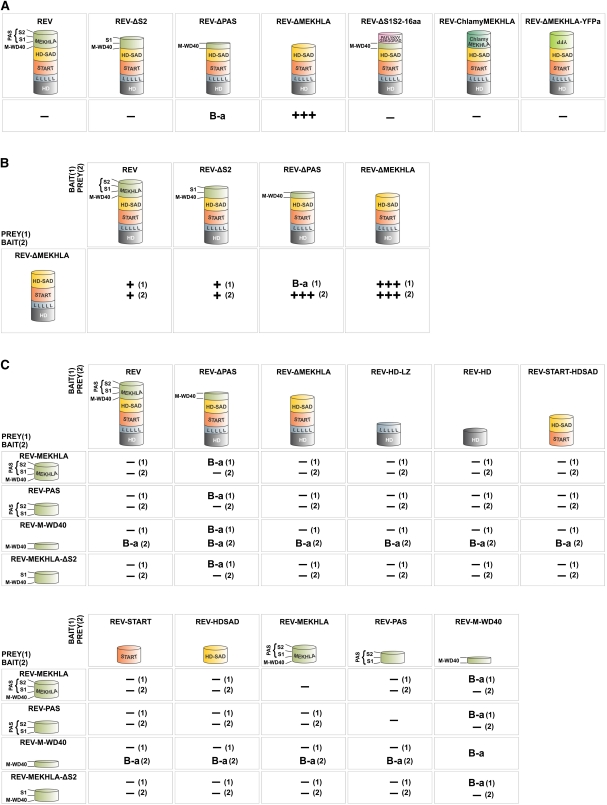
Figure 3.
The REV MEKHLA Domain Sterically Inhibits REV Homodimerization in Yeast.
Strong, weak, and no interactions are indicated by three plus signs, one plus sign, and a minus sign, respectively. Interaction strengths were determined by comparison to a set of standards (see Supplemental Figure 5 online). Reciprocal bait/prey couples are indicated by a number in parenthesis. B-a, bait autoactivation.
(A) Homodimerization Y2H assays. The same proteins were used as bait (fused to GAL4 DNA binding domain) and prey (fused to GAL4 activation domain).
(B) Interaction of REV-ΔMEKHLA with REV truncated for subsections of the MEKHLA domain.
(C) Interaction of isolated MEKHLA variants with REV protein domains.
[See online article for color version of this figure.]
Homodimerization of HD-ZIP III transcription factors is necessary for DNA binding in vitro and therefore presumably required in vivo for REV function (Sessa et al., 1997, 1998). It is generally assumed that homodimerization requires the leucine zipper domain. To test this assumption in vivo, we assayed REV proteins in which the leucine zipper had been removed for their ability to dimerize. Consistent with a role for the leucine zipper in dimerization, we found that REV proteins lacking the leucine zipper (REV-ΔLZ and REV-ΔLZ-ΔMEKHLA) are unable to homodimerize in yeast (see Supplemental Figure 2A online). Furthermore, the overexpression of REV*-ΔLZ in Arabidopsis failed to generate any of the phenotypes seen in lines overexpressing full-length REV* (data not shown) and failed to activate pZPR3-reporter expression in a tobacco transient expression assay (see Supplemental Figure 2B online). REV*-ΔLZ protein stability was tested by transiently expressing the protein fused to yellow fluorescent protein (YFP) in tobacco (see Supplemental Figure 2C online).
To determine which region of the MEKHLA domain inhibits REV homodimerization in yeast, we performed Y2H analyses with REV truncated in various positions. The entire MEKHLA domain was not required to prevent dimerization; the M-WD40 plus S1 subdomains were sufficient (Figure 3A). We were not able to test whether the M-WD40 region alone is sufficient to prevent dimerization because REV proteins in which both S1 and S2 subdomains are removed showed substantial autoactivation when used as bait (i.e., fused to the GAL4 DNA binding domain) (Figure 3A). Thus, in yeast, the M-WD40 domain, in the absence of S1 and S2, confers transcriptional activation capability to the REV protein and to the GAL4 DNA binding domain alone. Addition of the S1 domain is sufficient to inhibit this activation capability (Figure 3A).
The presence of the MEKHLA domain on only one member of a pair of REV proteins is sufficient to inhibit dimerization (Figure 3B), albeit less strongly than when the MEKHLA domain is present on both members of a pair. Because we could use a REV-ΔMEKHLA protein as bait in this experiment, we were able to determine that the M-WD40 subdomain is not sufficient to prevent REV dimerization, at least when present on one member of a dimer (Figure 3B).
To better understand how the MEKHLA domain inhibits REV homodimerization, we conducted a reverse Y2H screening using REV-ΔMEKHLA prey against REV bait randomly mutagenized in the MEKHLA domain (REV-mMEKHLA). We then screened for mutations that that would allow an interaction of REV-ΔMEKHLA and REV-mMEKHLA comparable to REV-ΔMEKHLA homodimerization. We obtained 28 positive colonies: 26 carried premature stop codons and two carried deletions promptly followed by a stop codon (Figure 1A). All mutations are located in the first 56 amino acids of the REV MEKHLA domain that include the M-WD40 subdomain (Figure 1A) and end right before the S1 region. Our screen retrieved 16 out of 22 possible premature stop codons obtainable by point mutation in the first 56 amino acids of the domain (Figure 1A). By contrast, we did not detect any of the 32 possible stop codons scattered throughout the PAS region (Figure 1A. These results confirm that part of or all the S1 region of the MEKHLA domain is necessary to inhibit REV homodimerization in yeast.
In summary, the Y2H data demonstrate that the M-WD40 plus S1 region is sufficient to prevent REV homodimerization. A simple way that this could occur would be for the MEKHLA domain to interact intramolecularly with the leucine zipper domain, thereby making it inaccessible to a partner molecule. To determine if the MEKHLA domain has the capability to engage in any intramolecular interaction, we tested the MEKHLA, PAS (S1+S2), and M-WD40 regions against several truncations and independent domains of REV by Y2H. We did not detect any interaction (Figure 3C). Furthermore, the MEKHLA domain and PAS subdomain did not inhibit REV-ΔMEKHLA homodimerization when expressed in trans in a yeast three-hybrid experiment (data not shown). Therefore, our results suggest that the MEKHLA domain does not act by binding intramolecularly to other REV domains.
An alternative possibility is that the MEKHLA domain interacts with an as yet unknown protein, thus sequestering the REV protein and inhibiting dimerization. This mechanism would predict the requirement of a specific amino acid sequence within the MEKHLA domain for such an interaction to occur. However, we find that when we replace the REV MEKHLA domain with a random peptide of similar length (REV-ΔMEKHLA-YFPa, where YFPa is the reverse complement of the YFP sequence), the random protein sequences prevent dimerization (Figure 3A). Similarly, if we replaced the S1+S2 region with a random 16-mer peptide (REV-ΔS1S2-16aa), this sequence was also able to prevent dimerization (Figure 3A). The simplest explanation for this result is that REV adopts a conformation such that the MEKHLA domain occupies space that prevents REV monomers from coming in contact with one another.
The MEKHLA Domain Modulates REV Dimerization in the Plant
To determine the role of the MEKHLA domain in regulating dimerization in the plant, we performed transient bimolecular fluorescence complementation (BiFC) assays in tobacco using the same REV truncation and fusion proteins described above. In BiFC experiments, the N-terminal portion of YFP is fused to one of the putative interaction partners and the C-terminal portion of YFP is fused to the other partner. Interaction between the two partners brings the two proteins close enough together that the YFP protein is reconstituted and can fluoresce (Hu et al., 2002). In this experiment, the mRNAs were made miRNA resistant through a silent mutation in the miRNA binding site (constructs carrying mutations that render REV miRNA resistant are designated REV*). In addition, we fused all REV* protein forms to YFP as a control for protein stability. In all cases, the fusion proteins were stable (Figure 4).
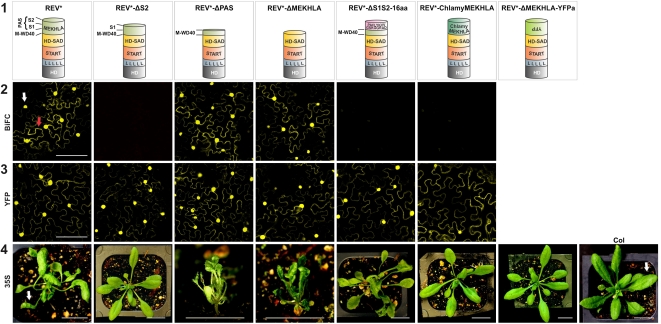
Figure 4.
The REV S1 Subdomain Is Sufficient to Inhibit REV Homodimerization, While the PAS Region Is Necessary for Relief of Inhibition.
Row 1: Schematic representation of the REV protein forms tested in the experiments below. miRNA-resistant REV is designated REV*. Row 2: BiFC assays in tobacco leaf abaxial (lower) epidermal cells transiently transformed. Nuclear and cytoplasmatic expression is indicated by a white and red arrow, respectively. Bar = 100 μm. Row 3: YFP fusion assays in tobacco leaf abaxial (lower) epidermal cells transiently transformed. Bar = 100 μm. Row 4: Overexpression analyses in stably transformed Arabidopsis plants. Representative individuals of 6-week-old transgenic plants are shown. White arrows point to a normally flattened leaf (right panel) and to an abnormally curled up leaf (left panel). Bar = 100 mm.
Similar to the Y2H data, the presence of the M-WD40 and the S1 subdomains together (REV*-ΔS2) prevented dimerization in vivo (Figure 4). Also, similar to the situation in yeast, replacing the PAS domain with a random 16–amino acid sequence prevented dimerization in the BiFC assay.
In contrast with the results in the Y2H experiment, full-length REV* and REV*-ΔMEKHLA dimerized equally well in BiFC experiments (Figure 4). To explain this apparent contradiction, we hypothesize that the S2 domain, in the context of a cellular signal, is capable of relieving inhibition by the M-WD40-S1 region and that the cellular context within which the transgenes are expressed (abaxial tobacco leaf epidermal cells) is one in which this signal is active.
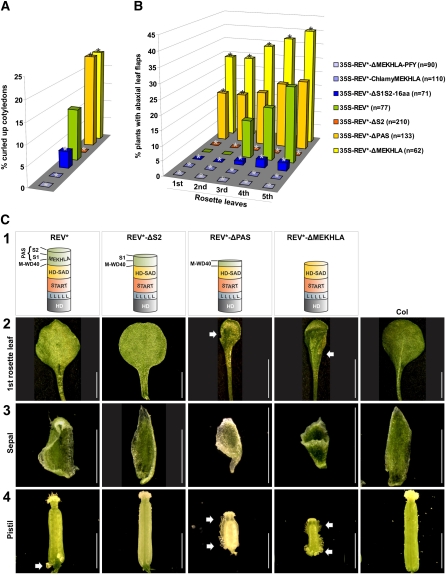
When overexpressed in plants, the REV* constructs truncated for different portions of the MEKHLA domain showed various behaviors. T1 transgenic plants were assayed for the shape of their cotyledons (ectopic REV activity causes the production of inwardly curled, adaxialized cotyledons) and for the presence of leaf flaps on the abaxial (lower) leaf surfaces. Leaf flaps occur when small regions of adaxial cell fate, caused by ectopic HD-ZIP III activity, are present on the bottom of the leaf.
Compared with a full-length REV* construct, which causes production of leaf flaps on leaves beginning with the third rosette leaf, REV* lacking a MEKHLA domain causes the production of such flaps earlier in development, beginning with the first two leaves, and on a higher proportion of leaves (Figures 4 and 5B). REV* lacking a MEKHLA domain also increases the fraction of cotyledons that are curled inward (Figure 5A). Effects on the morphologies of floral organs were also observed. REV*-ΔMEKHLA caused the production of shorter carpels with ectopic (i.e., growing out from the abaxial [outer] rather than adaxial [inner] carpel domain) ovules forming along the length of the carpel. This is in contrast with full-length REV*, which causes production of ectopic (abaxial) ovules only at the base of the carpel (Figure 5C). Sepals of plants overexpressing REV*-ΔMEKHLA were observed to be trumpet shaped, while this was not observed in plants overexpressing full-length REV* (Figure 5C). Thus, REV* lacking a MEKHLA domain acts earlier and more strongly in generating ectopic adaxial fates than a full-length REV* protein. This is consistent with a role for MEKHLA as a negative regulator.
Figure 5.
Loss of the REV MEKHLA Domain Results in More Severe Overexpression Phenotypes.
(A) Phenotypic analysis of stably transformed Arabidopsis plants. T1 seedlings were scored for curled up cotyledons caused by ectopic REV activity. miRNA-resistant REV is designated REV*. Asterisks indicate transgenic lines that are statistically different from 35S-REV* plants (Z test, confidence > 95%).
(B) Phenotypic analysis of stably transformed Arabidopsis plants. First to fifth rosette leaves of T1 seedlings were scored for abaxial (lower) leaf flaps. miRNA-resistant REV is designated REV*. Asterisks indicate transgenic lines that are statistically different from 35S-REV* plants (Z test, confidence > 95%).
(C) Row 1: Schematic representation of the REV protein forms overexpressed in the tissue samples below. Row 2: Abaxial side of the first rosette leaf of representative transgenic Arabidopsis plants. The arrows point to leaf flaps. Bar = 2 mm. Row 3: Sepals of representative transgenic Arabidopsis plants. Bar = 2 mm. Row 4: Pistils of representative transgenic Arabidopsis plants. The arrows point to abaxial ovules. Bar = 2 mm.
Overexpression of a construct in which the S2 domain alone is deleted, and in which we therefore predict the inhibition of dimerization cannot be overcome, does not cause any discernible overexpression phenotype (Figures 4 and 5) While it is possible that the REV-ΔS2 protein is unstable, our control BiFC experiments show that an N-terminal YFP fusion to the REV-ΔS2 protein is stable (Figure 4).
Overexpression of a REV protein in which only the PAS domain (S1+S2) is removed causes a phenotype similar to that in which the entire MEKHLA domain is removed (Figure 5). The effect on cotyledons is similar in magnitude to that seen in the REV*-ΔMEKHLA transgenic seedlings. The effect on the production of leaf flaps is similar in that it causes the production of these from an earlier stage, but the effect is not as dramatic in magnitude. The carpels also produce ectopic ovules along their entire lengths and sepals can be trumpet shaped. This is consistent with our observations that proteins lacking the PAS domain are able to undergo facile dimerization.
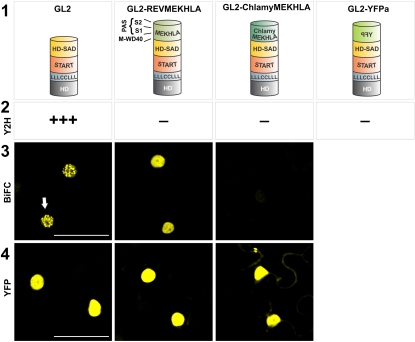
Unlike the class III HD-ZIP proteins, the class IV proteins do not carry a MEKHLA domain at their C termini. (They do, however, include the HD-ZIP, START, and HD-SAD domains.) To determine if the MEKHLA domain could prevent dimerization of the class IV HD-ZIP protein GLABRA2 (GL2), we fused the REV MEKHLA domain onto the GL2 protein (GL2-REVMEKHLA). Similar to what we observed with REV, the MEKHLA domain inhibited GL2 homodimerization in Y2H experiments but not in BiFC assays in planta (Figure 6). This inhibition was not sequence specific as, similar to the case for REV, replacing the MEKHLA domain with other sequences could prevent dimerization both in Y2H and BiFC assays. Thus, the class IV HD-ZIP proteins are similar to the MEKHLA-less HD-ZIPIII proteins in that their dimerization is sensitive to the presence of an extension at the C terminus and in that this sensitivity is not sequence specific. This is consistent with the observation that the class III and class IV HD-ZIP proteins have similar domain structures.
Figure 6.
The REV MEKHLA Domain Inhibits Homodimerization of the GL2 Protein.
Row 1: Schematic representation of the GL2 protein forms used in the experiments below. Row 2: Results of Y2H experiments. Strong and no interactions are indicated by three plus signs and a minus sign, respectively. Interaction strengths were determined by comparison to a set of standards (see Supplemental Figure 5 online). Row 3: BiFC assays in tobacco leaf abaxial (lower) epidermal cells transiently transformed. Nuclear expression is indicated by a white arrow. Row 4: YFP fusion assays in tobacco leaf abaxial epidermal cells transiently transformed. Bar = 100 μm.
[See online article for color version of this figure.]
We also investigated whether the C. reinhardtii MEKHLA domain could function to replace the REV MEKHLA domain and whether it could inhibit dimerization when fused to the class IV HD-ZIP protein GL2. Replacement of the REV MEKHLA domain with the C. reinhardtii MEKHLA domain resulted in inhibition of homodimerization in yeast (Figure 6) and also failure to form homodimers in BiFC assays. Similarly, the C. reinhardtii MEKHLA domain inhibited dimerization in both yeast and plant cells when fused to GL2. We hypothesize that the C. reinhardtii MEKHLA domain is not able to respond to cellular signals present in Arabidopsis to lift inhibition by the M-WD40+S1 region. We note that both the S1 and S2 domains are poorly conserved at the amino acid sequence level between the C. reinhardtii MEKHLA domain and the REV MEKHLA domain. Thus, these sequences may not be able to sense signals present in Arabidopsis that are responsible for lifting inhibition of dimerization by the MEKHLA domain.
The MEKHLA Domain of Other HD-ZIP III Proteins Behave Differently
The MEKHLA domain is conserved among all five Arabidopsis HD-ZIP III proteins (Figure 1). We performed Y2H dimerization assays with PHB and ATHB8, members of the other two HD-ZIP III phylogenetic clades (Figure 1). Both proteins behaved differently from REV as they strongly activated transcription as bait even in the absence of the MEKHLA domain (PHB-ΔMEKHLA and ATHB8-ΔMEKHLA) (Figure 7A). To avoid bait autoactivation, we tested PHB, PHB-ΔMEKHLA, ATHB8, and ATHB8-ΔMEKHLA as prey against REV and REV-ΔMEKHLA bait. PHB and ATHB8 interacted with REV-ΔMEKHLA more tightly than PHB-ΔMEKHLA and ATHB8-ΔMEKHLA did with REV (Figure 7A), suggesting that the REV MEKHLA domain has a stronger inhibitory effect than the MEKHLA domains of ATHB8 or PHB.
Figure 7.
The REV MEKHLA Domain Behaves Distinctly from the Other HD-ZIP III MEKHLA Domains.
(A) Results of Y2H experiments. Strong, medium, mild, and no interactions are indicated by three plus signs, two plus signs, one plus sign, and a minus sign, respectively. Interaction strengths were determined by comparison to a set of standards (see Supplemental Figure 5 online). Reciprocal bait/prey couples are indicated by a number in parenthesis. B-a, bait autoactivation.
(B) Results of Y2H experiments. No interaction is indicated by a minus sign.
(C) A representative individual of 35S-PHB* transgenic plants (4 weeks old). miRNA-resistant PHB is designated PHB*. Bar = 100 mm.
(D) A representative individual of 35S-PHB*-ΔMEKHLA transgenic plants (4 weeks old). miRNA-resistant PHB is designated PHB*. Bar = 100 mm.
In addition to forming homodimers, HD-ZIP III proteins form dimers with the small ZPR proteins (Wenkel et al., 2007; Kim et al., 2008). ZPR proteins consist almost entirely of a stretch of leucine zipper that is similar to the leucine zipper found in the class III HD-ZIP proteins. We assayed HD-ZIP III prey against ZPR3 bait. PHB and ATHB8, but not REV, strongly interacted with ZPR3 (Figure 7A). By contrast, PHB-ΔMEKHLA, ATHB8-ΔMEKHLA, and REV-ΔMEKHLA interacted equally well with ZPRIII (Figure 7A). These results show that REV and the other HD-ZIP III proteins have diverged from each other with REV possessing a unique way to modulate homo- and heterodimerization through the MEKHLA domain.
The REV MEKHLA domain is characterized by a stretch of four amino acids (AASE) that is not present in any other Arabidopsis HD-ZIP III protein (Figure 1A). These residues are not responsible for the unique activity of the REV MEKHLA domain observed in yeast as REV deleted for this peptide (REV-ΔAASE) did not homodimerize in Y2H assays (Figure 7B). Furthermore, we replaced the REV MEKHLA domain with the PHB MEKHLA domain to create REV-PHBMEKHLA. REV-PHBMEKHLA showed mild autoactivation as bait; no additional activity was observed when tested for homodimerization, indicating that REV-PHBMEKHLA does not homodimerize in yeast. This result suggests that the difference between REV and PHB homodimerization resides outside the MEKHLA domain (Figure 7B).
To test if the REV and PHB MEKHLA domains behave differently also in planta, we overexpressed a miRNA-resistant version of PHB and PHB-ΔMEKHLA (35S-PHB* and 35S-PHB*-ΔMEKHLA, respectively) in Arabidopsis. 35S-PHB* and 35S-PHB*-ΔMEKHLA lines showed indistinguishable overexpression phenotypes (Figures 7C and 7D), indicating that the PHB MEKHLA domain does not exert the same inhibitory effects observed with REV. One hypothesis is that REV plays unique roles in plant development that require dimerization regulation through the MEKHLA domain. In line with this model, REV is the only HD-ZIP III gene to show a loss-of-function phenotype. PHB and ATHB8 MEKHLA domains might carry out other functions. Alternatively, our experimental conditions did not allow us to detect the effect of the MEKHLA domain on PHB and ATHB8 dimerization.
REV Is the Latest Type of HD-ZIP III Protein to Have Evolved in Land Plants
To study the evolution of the MEKHLA domain we built a neighbor-joining tree based on the multiple sequence alignment of MEKHLA domains from key species in the evolution of land plants (Figure 1C; see Supplemental Figure 3 and Supplemental Data Set 1 online). To make the analysis more robust, we generated a neighbor-joining and a maximum parsimony tree with HD-ZIP III full-length sequences when available (Figure 1C; see Supplemental Figure 4 and Supplemental Data Set 2 online). The trees identify two major clades: one containing a more ancestral MEKHLA domain present in green algae, mosses, and ferns and the other containing a more recent MEKHLA domain present in angiosperms. Interestingly, gymnosperms possess both forms of MEKHLA domains. The trees could not fully resolve the evolution of angiosperm and more recent gymnosperm HD-ZIP III proteins. Nevertheless, the trees based on full-length sequences divided angiosperm HD-ZIP III proteins into two major phylogenetic clades: REV-PHB-PHV-like and ICU4/CNA-ATHB8. This phylogenetic analysis together with our functional data indicate that REV diverged considerably from the PHV/PHB and ICU4/CNA-ATHB8 clades and thus likely also from the more ancestral HD-ZIP III proteins. Therefore, we suggest that REV is the latest type of HD-ZIP III protein to have evolved in land plants.
DISCUSSION
Class III HD-ZIP proteins are master regulators of polar development in plants. They play important, conserved roles in the development of all parts of the plant: embryo, meristem, stem, leaf vasculature, root, and flower. The predicted structure of these transcription factors (the presence of a ligand binding domain and a PAS-like MEKHLA domain) indicate that there is substantial posttranslational regulation of HD-ZIP III function. In this article, we explored the function of the MEKHLA domain of HD-ZIP III REV. Our results indicate that this domain plays a negative regulatory role and support a model in which steric inhibition by the MEKHLA domain prevents dimerization of REV monomers. We speculate that a cellular signal recognizes the MEKHLA domain and changes its conformation to allow dimerization.
MEKHLA as a Negative Regulatory Domain That Prevents Dimerization
We assayed the ability of the REV protein to function with and without the MEKHLA domain in several ways; in each test, the MEKHLA-less protein was able to supply REV function. In some cases, the MEKHLA-less protein caused a more severe gain-of-function phenotype than the full-length protein. These observations all indicate that the MEKHLA domain plays a negative regulatory role. An important corollary of this is that MEKHLA negative regulation itself must be modulated to allow REV to be active at the correct time and location.
Only one point mutation has been described in the MEKHLA domain to date. This is a mutation in ICU4/CNA (Duclercq et al., 2010). A mutation at position 16 of the MEKHLA domain (this is in the beginning of the M-WD40 portion of the domain) changes a conserved Ser into a Cys in the hoc mutant. The hoc mutants were isolated in a screen for mutants that could regenerate shoots from explants in the absence of exogenous hormones (Catterou et al., 2002). The hoc mutation appears to be recessive. However, the mutation does not appear to cause a loss of protein function since mutations that disrupt the gene more severely fail to generate shoots on this medium. One interpretation of these observations is that repression by the MEKHLA domain is lifted by exogenous hormones in culture. In the hoc mutant, MEKHLA repression would be compromised and there would be no need for exogenous hormones.
Our experiments in yeast show that the presence of a MEKHLA domain on a REV protein prevents dimerization: when removed, REV dimerizes readily in yeast. We also find that the MEKHLA domain, when added to the HD-ZIP IV protein GL2, can inhibit its dimerization. The structure of class IV proteins is similar to that of class III proteins with the exception of the presence of the MEKHLA domain. Only a portion of the MEKHLA domain was needed; the M-WD40+S1 region is sufficient to carry out the inhibition. The S1 domain is critical, and the M-WD40 domain alone could not inhibit dimerization.
Interestingly, this effect was not sequence specific. The S1 domain could be replaced with random sequences, and this could inhibit REV homodimerization. In fact, the entire MEKHLA domain could be replaced with random sequences, and this prevented homodimerization in yeast. This finding would appear to rule out two possible mechanisms of MEKHLA action. In the first, the MEKHLA domain might interact intramolecularly with other regions of REV, for instance, the leucine zipper domain, and in so doing could prevent it from interacting with other proteins. This would require sequence specificity. In addition to the above results, we did not find any evidence that MEKHLA interacts with other REV domains despite extensive testing.
In a second possible mechanism, the MEKHLA domain could interact with an as yet unknown partner that acts to sequester the REV protein, thus preventing it from dimerizing. Again, this would require sequence specificity, and this does not appear to be the case. Also, yeast two-hybrid screens done in our laboratory for MEKHLA-dependent interactors failed to turn up any of these (our unpublished data). HD-ZIP III proteins have been reported to interact with the APETALA2-like transcription factors DORNROSCHEN and DORNROSCHEN-like (DRNL) through the MEKHLA domain (Chandler et al., 2007). This would be evidence for a sequence-specific interaction. However, in apparent contradiction to these results, we did not detect any interaction of DRNL with full-length REV or with the REV MEKHLA domain by Y2H. We do not know why our results differ from previous results. Furthermore, DRNL did not allow REV homodimerization when coexpressed in yeast with REV prey and REV bait. Therefore, we believe DRNL is likely not responsible for lifting the MEKHLA domain inhibition and might play a role in the function of other HD-ZIP III proteins.
We are left then with a third mechanism in which the MEKHLA domain sterically inhibits dimerization, perhaps by blocking access to the leucine zipper domain. The leucine zipper domain is believed to be the site of dimerization, and our in vivo studies are consistent with this. Such a steric masking mechanism has been reported to regulate endoplasmic reticulum retention of certain plasma membrane–bound receptors (Letourneur et al., 1995).
Our results from dimerization assays in plant cells (BiFC assays) differ from those obtained in yeast cells. Similar to the yeast assays, we find that the substitution of random sequences in place of the MEKHLA domain can inhibit dimerization of REV as well as GL2. We also find that the M-WD40 plus S1 subdomains can inhibit dimerization as they do in yeast. However, full-length MEKHLA domains allow dimerization. We hypothesize that this is because the full-length MEKHLA domain is able to receive a cellular signal that alters its conformation, moving it out of the way and allowing dimerization. This would not be possible for the random sequences. The S2 subdomain appears to be required for lifting this repression by the MEKHLA domain since the REV protein carrying only the M-WD40+S1 region, without the S2 subdomain, is not constitutively active and does not cause gain-of-function phenotypes. This is consistent with their ability to prevent dimerization. Interestingly, the C. reinhardtii MEKHLA domain prevents homodimerization both in yeast and in the BiFC assay. We propose that this distantly related protein is not able to respond to a cellular signal present in tobacco cells. We note that the S1 and S2 subdomains of the C. reinhardtii MEKHLA domain are quite diverged from that of the class III HD-ZIP proteins. This may indicate that the C. reinhardtii MEKHLA domains respond to other stimuli.
Different parts of the Arabidopsis plant responded differently to the overexpression of full-length or MEKHLA-less REV. Certain cell types, such as the abaxial (lower) cells of the first two rosette leaves and the upper part of the carpel, were not responsive to REV carrying a MEKHLA domain. We speculate that the putative signal responsible for relieving the MEKHLA inhibition is absent in such cell types. Carrying this logic further, cells that show a gain-of-function phenotype in response to full-length REV would be predicted to have high levels of such signals.
Much is known about the structure of PAS domains (Möglich et al., 2009). Most if not all of this work has been done on the PAS domain alone. PAS domains have been shown to move relative to one another in response to a stimulus (Möglich et al., 2009). Less is known about how the PAS domain functions when it is part of a larger protein with other domains. It is possible that a stimulus could cause movement of the PAS domain in HD-ZIP III proteins relative to other portions of the protein. Such a mechanism could work to relieve the steric masking we propose.
Evolution of the REV MEKHLA Domain
Algae and bacteria have single-domain MEKHLA proteins but not HD-ZIP proteins. By contrast, HD-ZIP III proteins are the only ones to carry a MEKHLA domain in land plants. A likely scenario is that HD-ZIP III proteins acquired a MEKHLA domain from an ancestral MEKHLA domain protein similar to those present in algae. Nevertheless, the C. reinhardtii MEKHLA domain could not effectively substitute for the REV MEKHLA domain, suggesting functional divergence of the two MEKHLA domains. The C. reinhardtii MEKHLA domain inhibited REV homodimerization both in yeast and in planta, indicating that it is not recognized by the putative cellular signal responsible for inhibition relief in planta. The REV and C. reinhardtii MEKHLA domains are highly divergent in sequence and might have evolved to recognize different signals. In line with this model, our phylogenetic tree separates the MEKHLA domains of angiosperms from those of more basal plants.
The REV MEKHLA domain maintained its function when fused to GL2, a class IV HD-ZIP protein that naturally does not carry a MEKHLA domain. This result suggests that HD-ZIP proteins that do not carry a MEKHLA domain are perfectly suited to accept one, in line with the domain acquisition model. Therefore, we speculate that class III HD-ZIP proteins evolved from class IV-like HD-ZIP proteins by acquiring a MEKHLA domain.
The MEKHLA domain is conserved among all HD-ZIP III proteins, with striking sequence similarity. Nevertheless, the MEKHLA domain of REV but not that of PHB or ATHB8 inhibited heterodimerization among HD-ZIP III proteins and inhibited interaction with ZPR3 proteins. Such a difference might lie not only in the MEKHLA domain but also in the rest of the protein, as a chimeric REV protein carrying the PHB MEKHLA domain did not homodimerize in yeast. Furthermore, the overexpression of full-length PHB or PHB deleted in the MEKHLA domain resulted in indistinguishable phenotypes, in contrast with what was observed with REV. These data indicate that the function and/or regulation of the REV MEKHLA domain are substantially different from that of other HD-ZIP III proteins. In line with this model, REV is the only HD-ZIP III to show a loss-of-function phenotype. PHB and ATHB8 MEKHLA domains might carry out completely different functions or might simply recognize a more ubiquitous signal that did not allow us to highlight functional differences in the condition tested. REV is expressed in a broader domain in the developing leaf than PHB or ATHB8 are (Prigge et al., 2005). This could be the basis of a requirement for a different type of negative regulation.
Our phylogenetic trees, based on the alignment of full-length HD-ZIP III proteins, group PHB, PHV, and REV in a separate phylogenetic clade from ICU4/CNA and ATHB8. Nevertheless, our functional assays showed that PHB behaves more similarly to ATHB8 than to REV. Together, these results suggest that REV diverged from the other HD-ZIP III proteins. Note that in some cases, PHB behaves genetically more similarly to ICU4/CNA than it does to REV (Prigge et al., 2005). It is possible that functional differences in the MEKHLA domain are responsible for the observed genetic and phenotypic differences seen in combinations of loss-of-function mutants.
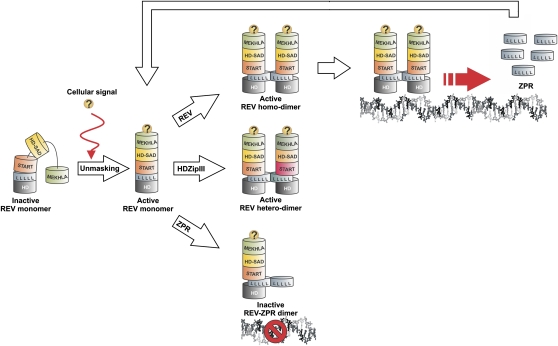
Model
Figure 8 shows a model for regulation of REV dimerization and function. In the absence of a repression lifting signal (indicated with a question mark), the MEKHLA domain obstructs access to the leucine zipper domain of REV, preventing interaction with either other HD-ZIP III proteins or with the ZPR proteins. In response to the as yet unknown repression lifting signal, REV may dimerize either with itself (active form), with other HD-ZIP III proteins (presumed active form), or with ZPR proteins (inactive or altered activity form). When active, REV stimulates transcription of the ZPR proteins, which feedback to limit REV activity.
Figure 8.
Model for Regulation of REV Dimerization by the MEKHLA Domain.
The MEKHLA domain prevents REV dimerization in a steric fashion. An unknown mechanism would then recognize the sequence of the MEKHLA domain and relieve the inhibition. This molecular switch precedes REV homodimerization and heterodimerization with other HD-ZIP III proteins and interaction with ZPR proteins.
[See online article for color version of this figure.]
We speculate that an ancestral MEKHLA protein fused onto a class IV–like HD-ZIP protein to give rise to class III HD-ZIP proteins. The REV MEKHLA domain might have diverged to acquire functions separate from other HD-ZIP III proteins.
METHODS
Bioinformatics Techniques
Multiple sequence alignments were constructed using Muscle 3.2 (Edgar, 2004) and displayed with Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) using the BLOSUM62 matrix and a 50% threshold for shading. Protein secondary structure was predicted using the JPRED3 (Cole et al., 2008). Phylogenetic trees were constructed using the neighbor-joining or maximum parsimony algorithm implemented in the MEGA software suite (Tamura et al., 2007). We used mean character difference, among-site rate variation, and random seed initiation; 10,000 bootstrap replicates were performed followed by identification of the consensus tree.
Molecular Biology
The expression level of transgenes was checked by RT-PCR analyses as described by Sambrook and Russell (2001). PCR analyses were conducted using the REV-F and REV-R4 primers (REV-F, 5′-ATGGAGATGGCGGTGGCTAAC-3′; REV-R4, 5′-CCTCGACGCCTCTTTCCTCTGC-3′), and the amplification products were visualized with ethidium bromide. β-Glucuronidase activity was tested as described by Jefferson (1989).
Cloning
Coding sequences (see Supplemental Table 1 online) were PCR amplified (with a start codon and a stop codon when missing) and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer's instructions. PCR primers were all designed as 25-bp oligomers starting and ending at the locations indicated in Supplemental Table 1 online. The miRNA-resistant rev and phb mutations were previously described (Mallory et al., 2004; Wenkel et al., 2007).
The REV-ΔMEKHLA and REV-ΔS1S2 coding sequences were PCR amplified with a BamHI restriction site at the 5′ end and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer's instructions to create REV-ΔMEKHLA-BamHI and REV-ΔS1S2-16aa, respectively. The sequence coding for the Chlamydomonas reinhardtii MEKHLA domain and the YFP reverse-complement sequence were PCR amplified with a start codon and a stop codon flanked by BglII restriction sites and cloned into the BamHI restriction site of REV-ΔMEKHLA-BamHI to create REV-ChlamyMEKHLA and REV-YFPa, respectively. REV-ΔAASE and REV-PHBMEKHLA were obtained by overlap extension PCR. The GL2 coding sequence was PCR amplified with a BamHI restriction site at the 5′ end and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer's instructions to create GL2-BamHI. The sequence coding for the C. reinhardtii MEKHLA domain, the REV-MEKHLA sequence, and the YFP reverse complement sequence were PCR amplified with a start codon and a stop codon flanked by BglII restriction sites and cloned into the BamHI restriction site of GL2-BamHI to create GL2-ChlamyMEKHLA, GL2-REV-MEKHLA, and REV-YFPa, respectively.
For Y2H assays, sequences were mobilized from pENTR/D-TOPO into pDEST22 (Invitrogen) and pDEST32 (Invitrogen) according to the manufacturer's instructions.
For overexpression assays, sequences were mobilized from pENTR/D-TOPO into pMDC32 (Curtis and Grossniklaus, 2003).
For BiFC assays, the sequences coding for the N terminus and C terminus of YFP were PCR amplified from pSPYNE-35S and pSPYCE-35S (Walter et al., 2004) using primers carrying KpnI and AscI restriction sites and cloned into the KpnI and AscI restriction sites of pMDC32 to create pMDC32-SPYNE and pMDC32-SPYCE, respectively. Sequences were mobilized from pENTR/D-TOPO into pMDC32-SPYNE and pMDC32-SPYCE according to the manufacturer's instructions.
For YFP fusion analyses, sequences were mobilized from pENTR/D-TOPO into pEarleyGate-104 (Earley et al., 2006) according to the manufacturer's instructions.
Y2H Assays
The Invitrogen ProQuest two-hybrid system with Gateway Technology was used in Y2H assays according to the manufacturer's instructions. Positive interactions were detected by assaying for β-galactosidase activity at 8 and 24 h. More than six independent colonies per couple of constructs were tested. To assess the interaction strength of our protein couples, we used a collection of control strains, provided with the Invitrogen system, that contain plasmid pairs expressing fusion proteins with a spectrum of interaction strengths. +++, ++ and + correspond to interactions similar in strength to those observed with pPC97-Fos/pPC86-Jun, pPC97-CYH2–dDP/pPC86-dE2F, and pPC97-RB/pPC86-E2F1, respectively (see Supplemental Figure 5 online).
For the reverse Y2H screening, the sequence encoding the REV MEKHLA domain was mutagenized by PCR with Mutazyme (Stratagene). The PCR products were then cotransformed with pDEST32-REV, linearized in the XbaI restriction site, into yeast cells carrying pDEST22-REV-ΔMEKHLA.
Plant and Genetic Materials
Seeds of Arabidopsis thaliana, ecotype Columbia-0 or Nossen-0, were used for all experiments. The rev-1 mutant was previously described (Talbert et al., 1995; Ratcliffe et al., 2000). Plants were grown in the greenhouse under long-day conditions or in vitro under constant fluorescent illumination.
Transgenic Plants
For stable transformation, Agrobacterium tumefaciens strain GV3101 was used to transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Expression levels were analyzed by RT-PCR. For transient transformation, Nicotiana benthamiana abaxial epidermal cells were infiltrated with Agrobacterium strain GV3101 as previously described (Neuhaus and Boevink, 2001).
Microscopy
BiFC and YFP fusion experiments were conducted by infiltrating tobacco leaves with Agrobacterium strains carrying the appropriate fusion vectors. For each experiment, four different construct combinations were infiltrated into the same leaf. Each experiment was repeated three times. The REV constructs (either pSPYNE-35S-REV*/pSPYCE-35S-REV* or pEarleyGate-104-REV*) were infiltrated every time as reference standards for BiFC and YFP fusion experiments, respectively. YFP visualizations in tobacco epidermal cells were performed 40 to 48 h after infiltration using a Leica SP5 confocal laser scanning microscope. For each experiment, the same gain setting was used to visualize the cells infiltrated with the control constructs and the cells infiltrated with the experimental constructs.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: REV (At5g60690), PHB (At2g34710), ATHB8 (At4g32880), GL2 (At1g79840), ZPR3 (At3g52770), and C. reinhardtii gene encoding a MEKHLA domain (XM_001691785).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The REVOLUTA M-WD40 Subdomain Shows Similarities to the WD40 Domain.
Supplemental Figure 2. The Leucine Zipper Domain Is Necessary for REVOLUTA Transcriptional Activation Activity in Vivo.
Supplemental Figure 3. Multiple Sequence Alignment of the Arabidopsis, Oryza sativa, Ginkgo biloba, Pinus taeda, Psilotum nudum, Physcomitrella patens, and Chlamydomonas reinhardtii MEKHLA Domains Used to Generate the Neighbor-Joining Tree in Figure 1C.
Supplemental Figure 4. Multiple Sequence Alignment of the Arabidopsis, O. sativa, G. biloba, P. taeda, P. nudum, and P. patens HD-ZIP III Proteins Used to Generate the Neighbor-Joining and Maximum-Parsimony Trees Built to Corroborate the Tree in Figure 1C.
Supplemental Figure 5. Yeast Two-Hybrid Assays.
Supplemental Table 1. Sequences Used in This Study.
Supplemental Data Set 1. Amino Acid Sequences Used to Generate the Alignment Presented in Supplemental Figure 3 online.
Supplemental Data Set 2. Amino Acid Sequences Used to Generate the Alignment Presented in Supplemental Figure 4 online.
Acknowledgments
We thank the members of the Barton lab for valuable discussions and technical advice. The work was supported by a grant from the National Science Foundation (to M.K.B.). E.M. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
References
- Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baima S., Possenti M., Matteucci A., Wisman E., Altamura M.M., Ruberti I., Morelli G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou M., Dubois F., Smets R., Vaniet S., Kichey T., Van Onckelen H., Sangwan-Norreel B.S., Sangwan R.S. (2002). hoc: An Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. Plant J. 30: 273–287 [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Grewe B., Werr W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36(Web Server issue): W197––W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J., Ndong Y.P.A., Guerineau F., Sangwan R.S., Catterou M. (2010). Arabidopsis shoot organogenesis is enhanced by an amino acid change in the ATHB15 transcription factor. Plant Biol. 13: 317–324 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Floyd S.K., Zalewski C.S., Bowman J.L. (2006). Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173: 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.A., Prigge M.J., Katzman R.B., Clark S.E. (2005). CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg S.P., Galinha C., Kornet N., Canales C., Scheres B., Tsiantis M. (2009). Repression of apical homeobox genes is required for embryonic root development in Arabidopsis. Curr. Biol. 19: 1485–1490 [DOI] [PubMed] [Google Scholar]
- Hawker N.P., Bowman J.L. (2004). Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Lee M., Lee I., Park H.Y., Seo P.J., Jung J.H., Kwon E.J., Suh S.W., Paek K.H., Park C.M. (2008). HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F., Hennecke S., Démollière C., Cosson P. (1995). Steric masking of a dilysine endoplasmic reticulum retention motif during assembly of the human high affinity receptor for immunoglobulin E. J. Cell Biol. 129: 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McHale N.A., Koning R.E. (2004). MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16: 1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Ayers R.A., Moffat K. (2009). Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17: 1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Bürglin T.R. (2006). MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 140: 1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus J.M., Boevink P. (2001). Plant Cell Biology. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Ochando I., González-Reig S., Ripoll J.J., Vera A., Martinez-Laborda A. (2008). Alteration of the shoot radial pattern in Arabidopsis thaliana by a gain-of-function allele of the class III HD-Zip gene INCURVATA4. Int. J. Dev. Biol. 52: 953–961 [DOI] [PubMed] [Google Scholar]
- Ochando I., Jover-Gil S., Ripoll J.J., Candela H., Vera A., Ponce M.R., Martínez-Laborda A., Micol J.L. (2006). Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol. 141: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Aravind L. (1999). START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Clark S.E. (2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8: 350–361 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C., Lackner P., Breiteneder H. (2008). The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Riechmann J.L., Zhang J.Z. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12: 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Ruberti I., Sessa G., Lucchetti S., Morelli G. (1991). A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 10: 1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K., Nishiyama T., Kato M., Hasebe M. (2001). Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol. Biol. Evol. 18: 491–502 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schena M., Davis R.W. (1992). HD-Zip proteins: Members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 89: 3894–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Morelli G., Ruberti I. (1993). The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12: 3507–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Morelli G., Ruberti I. (1997). DNA-binding specificity of the homeodomain-leucine zipper domain. J. Mol. Biol. 274: 303–309 [DOI] [PubMed] [Google Scholar]
- Sessa G., Steindler C., Morelli G., Ruberti I. (1998). The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Smith Z.R., Long J.A. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tron A.E., Welchen E., Gonzalez D.H. (2004). Engineering the loop region of a homeodomain-leucine zipper protein promotes efficient binding to a monomeric DNA binding site. Biochemistry 43: 15845–15851 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wenkel S., Emery J., Hou B.H., Evans M.M.S., Barton M.K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L., Grigg S.P., Xie M., Christensen S., Fletcher J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11: 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G.K., Kubo M., Zhong R., Demura T., Ye Z.H. (2007). Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 48: 391–404 [DOI] [PubMed] [Google Scholar]
- Zhulin I.B., Taylor B.L., Dixon R. (1997). PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22: 331–333 [DOI] [PubMed] [Google Scholar]