This study uses a combination of molecular approaches, in vivo imaging of fluorescent proteins, and structural analysis by electron tomography to study the synthesis and transport of storage proteins in aleurone cells. It describes an unusual autophagic mechanism for the delivery of storage proteins to the vacuole that may be common to cereals.
Abstract
Zeins, the prolamin storage proteins found in maize (Zea mays), accumulate in accretions called protein bodies inside the endoplasmic reticulum (ER) of starchy endosperm cells. We found that genes encoding zeins, α-globulin, and legumin-1 are transcribed not only in the starchy endosperm but also in aleurone cells. Unlike the starchy endosperm, aleurone cells accumulate these storage proteins inside protein storage vacuoles (PSVs) instead of the ER. Aleurone PSVs contain zein-rich protein inclusions, a matrix, and a large system of intravacuolar membranes. After being assembled in the ER, zeins are delivered to the aleurone PSVs in atypical prevacuolar compartments that seem to arise at least partially by autophagy and consist of multilayered membranes and engulfed cytoplasmic material. The zein-containing prevacuolar compartments are neither surrounded by a double membrane nor decorated by AUTOPHAGY RELATED8 protein, suggesting that they are not typical autophagosomes. The PSV matrix contains glycoproteins that are trafficked through a Golgi-multivesicular body (MVB) pathway. MVBs likely fuse with the multilayered, autophagic compartments before merging with the PSV. The presence of similar PSVs also containing prolamins and large systems of intravacuolar membranes in wheat (Triticum aestivum) and barley (Hordeum vulgare) starchy endosperm suggests that this trafficking mechanism may be common among cereals.
The cereal endosperm consists of three main cell types: an inner mass of starchy endosperm cells, one to three layers of epidermal aleurone cells, and the transfer cells that contact the maternal vascular tissue (Olsen, 2004). The starchy endosperm accounts for 80 to 90% of the grain weight and contains large amounts of storage proteins and starch. These cells undergo programmed cell death during maturation. Aleurone cells are rich in protein storage vacuoles (PSVs), minerals, and lipid bodies and remain alive during seed development. It is assumed that the breakdown of proteins localized to PSVs in aleurone cells provides an essential source of the amino acids necessary for the synthesis of hydrolytic enzymes required for mobilizing food stored in the starchy endosperm (Filner and Varner, 1967; Jacobsen et al., 1988; Bethke et al., 1998).
Besides its biological relevance as a model to study plant development, cell differentiation, and programmed cell death, the cereal endosperm is very important in terms of its nutritional value. Cereal grains contain less protein than do legume seeds, but because cereals are produced and consumed in much larger quantities, they are the main source of protein for the nutrition of humans and livestock (Shewry and Halford, 2002). The major storage proteins in maize (Zea mays) kernels are the alcohol-soluble prolamins, a type of storage proteins present only in grasses. Maize prolamins, referred to as zeins, are divided into different types: α-, β-, γ-, and δ-zeins (Coleman and Larkins, 1999) that differ in amino acid composition and structural properties (Shewry and Halford, 2002). Zeins are synthesized in the endoplasmic reticulum (ER) where they form accretions called ER protein bodies. Zeins and other prolamins do not contain a canonical ER retention signal, and the mechanism responsible for their aggregation involves specific protein–protein interactions among each other, the action of the chaperone binding protein (Bip), and the formation of disulfide bridges (Li et al., 1993; Coleman et al., 1996; Alvarez et al., 1998; Kim et al., 2002; Vitale and Ceriotti, 2004; Randall et al., 2005; Pompa and Vitale, 2006; Kumamaru et al., 2007). Maize seeds also contain smaller quantities of legumin-1 and α-globulin (Woo et al., 2001; Yamagata et al., 2003). These storage proteins accumulate in vacuoles during the early stages of endosperm development and are then retained in ER protein bodies at later stages (Arcalis et al., 2010).
In the starchy endosperm of wheat (Triticum aestivum) and barley (Hordeum vulgare), prolamins are also synthesized in the ER, but they are further transported to PSVs in a process that seems to involve Golgi-dependent and Golgi-independent pathways (Levanony et al., 1992; Galili et al., 1993; Rechinger et al., 1993). Autophagy and de novo formation of PSVs has also been reported to mediate the transport of prolamins to the PSVs in wheat (Levanony et al., 1992), but the molecular and cellular mechanisms underlying these routes remain unknown.
Autophagy involves the delivery of cytoplasmic components to the vacuole/lysosome and is conserved among eukaryotes (Thompson and Vierstra, 2005). The best-characterized autophagic process is macroautophagy, in which a reticulated cage-like structure called phagophore or preautophagosomal structure encircles the material to be degraded. The phagophore elongates and eventually closes around the engulfed cytoplasmic material, creating a sealed double membrane–bound compartment or autophagosome. The external membrane of the autophagosome fuses to the vacuole, thus releasing a single membrane vesicle or autophagic body that is degraded by acidic vacuolar hydrolases. Approximately 30 AUTOPHAGY-RELATED (ATG) proteins are involved in macroautophagy in yeast (Saccharomyces cerevisiae) (Geng and Klionsky, 2008; Behrends et al., 2010). One of them, ATG8, is a ubiquitin-like protein that becomes conjugated to phosphatidylethanolamine (PE) by the E1-like ATG7 and E2-like ATG3 proteins. Lipidated ATG8 becomes embedded in the phagophore and autophagosome membranes and probably acts as a scaffold to support cargo recruitment and membrane expansion (Geng and Klionsky, 2008). In fact, the amount of ATG8-PE positively correlates with the size of autophagosomes in yeast (Xie et al., 2008). In some cases, a second type of macroautophagy occurs by a mechanism independent of ATG8 lipidation (Nishida et al., 2009). Microautophagy is also possible; here, cytoplasmic material is directly engulfed by the vacuolar membrane (tonoplast), followed by release of the vesicle inside the vacuolar lumen (Bassham et al., 2006).
As described above, aleurone cells store proteins not in ER protein bodies but rather in PSVs. Although in situ hybridization studies suggest that zein genes are expressed in the maize aleurone layer (Woo et al., 2001), prolamins have not been reported to accumulate in cereal aleurone cells. There are no studies on what type of proteins may be stored in the maize aleurone PSVs or on the pathways involved in their vacuolar transport. Some of our limited knowledge is caused by the difficulty in preserving endosperm tissue for electron microscopy and by the lack of fluorescent subcellular markers in cereals.
In this study, we use a combination of molecular approaches, in vivo imaging of fluorescent proteins, and structural analysis by electron tomography of high-pressure frozen/freeze-substituted maize endosperms to study the synthesis and transport of storage proteins in aleurone cells. We show that zeins, α-globulin, and legumin-1 accumulate inside PSVs in maize aleurone cells. Their transport seems to require an atypical autophagic process that delivers ER material directly to PSVs by a process potentially independent of ATG8 lipidation. Prolamin-containing PSVs similar to the ones analyzed here have been reported in the wheat and barley starchy endosperms, suggesting that this may be a common mechanism for the vacuolar delivery of prolamins in cereals.
RESULTS
Zeins Are Expressed in Both Aleurone and Starchy Endosperm Cells
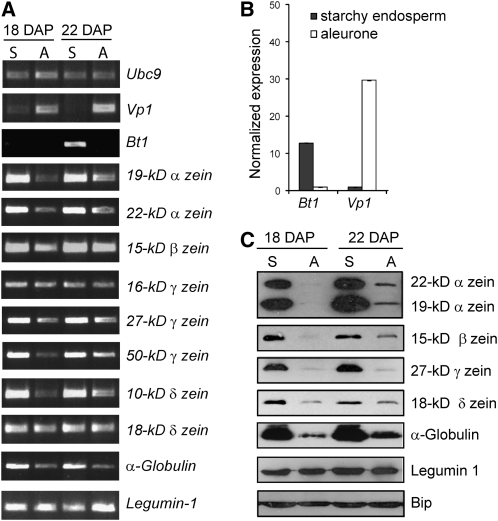
To determine whether storage protein genes, which are highly expressed in the maize starchy endosperm (Woo et al., 2001), are also expressed in aleurone cells, we examined by RT-PCR their expression in excised aleurone peels and starchy endosperm from maize B73-inbred kernels (see Supplemental Figure 1 online). To estimate cross-contamination between the two tissues, we extracted RNA and amplified transcripts of Brittle1 (Bt1) and Viviparous1 (Vp1), which are genes preferentially expressed in the starchy endosperm and aleurone cells, respectively (Shannon et al., 1998; Cao et al., 2007; Kirchberger et al., 2007). Quantification via real-time RT-PCR showed a 12.8-fold enrichment of Bt1 transcripts in starchy endosperm samples and 29.7-fold enrichment of Vp1 transcripts in aleurone samples, indicating very low cross-contamination between the two cell types (Figures 1A and 1B). When the same RNAs were used to amplify transcripts of storage proteins by RT-PCR, we detected transcripts for the 19-kD d2-α-, 22-kD z1-α-, 15-kD β-, 16-kD γ-, 27-kD γ-, 50-kD γ-, 10-kD δ-, and 18-kD δ-zeins, as well as α-globulin and legumin-1 in both tissue types (Figure 1A). These data confirmed the presence of zein transcripts in the aleurone layer as first proposed by Woo et al. (2001) using in situ hybridization.
Figure 1.
Expression of Zeins and Other Storage Proteins in Aleurone and Starchy Endosperm Cells.
(A) RT-PCR analysis of expression of genes encoding zeins, α-globulin, and legumin-1 in aleurone peels (A) and starchy endosperm samples (S) from 18- and 22-DAP kernels. Ubc9 (Ubiquitin-conjugating enzyme 9; Chung et al. 2009), Bt1 (starchy endosperm-preferred expression), and Vp1 (aleurone-preferred expression) were used as controls. PCR products were visualized by staining with ethidium bromide.
(B) Expression profile of Bt1 and Vp1 in aleurone and starchy endosperm samples from 18-DAP kernels determined by real-time quantitative RT-PCR. The expression of both genes in the two cell types was first normalized to Ubc9 and then normalized to the transcript levels of that particular gene in the starchy endosperm (Vp1) or in the aleurone cells (Bt1). The results represent the average and the standard error of two biological samples analyzed by triplicate.
(C) Immunoblot analysis of storage proteins in aleurone (A) and starchy endosperm (S) protein extracts. Bip was used as loading control.
Since zeins have not been reported previously to accumulate in aleurone tissue, we extracted proteins from aleurone peels and starchy endosperm tissue and performed immunoblot analysis to detect zeins and other storage proteins using specific antibodies (Woo et al., 2001). Interestingly, whereas the α-globulin, 22-kD α-, 19-kD α-, 15-kD β-, and 27-kD γ-zeins were abundant in the starchy endosperm, substantially reduced amounts of these polypeptides were detected in aleurone cells (Figure 1C). Not all storage proteins were comparably reduced, as the 18-kD δ-zein and legumin-1 were found at low but easily detectable levels in aleurone cells (Figure 1C). The detection of zein transcripts as well as zein proteins in the aleurone peels demonstrates that these storage proteins accumulate in aleurone cells.
Zeins Are Stored inside PSVs in Aleurone Cells
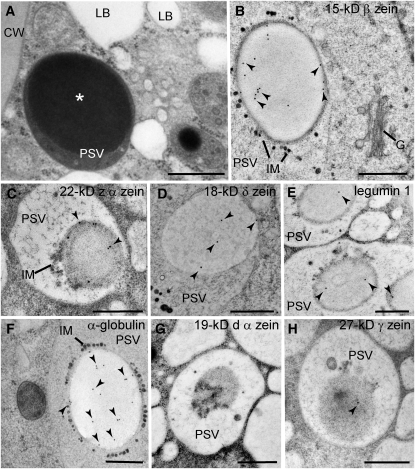
We then analyzed the subcellular localization of zeins, α-globulin, and legumin-1 in aleurone cells by immunogold labeling of high-pressure frozen/freeze-substituted B73 kernels at 18 and 22 d after pollination (DAP). Aleurone cells do not contain typical ER protein bodies, but rather have PSVs with large inclusions (Figure 2A). Whereas very weak or no signal was detected from the 19-kD α- and 27-kD γ-zeins in aleurone cells, we could detect the 22-kD α-, 15-kD β-, and 18-kD δ-zeins, as well as α-globulin and legumin-1 in PSVs (Figures 2B to 2H). More precisely, these storage proteins localized to large PSV inclusions (Figures 2B to 2H). By contrast, none of these storage proteins were detected on Golgi bodies (at least 10 Golgi stacks for each immunogold-labeling experiment were examined) or Golgi-associated vesicles (for example, Figure 2B). All the antibodies heavily labeled starchy endosperm protein bodies (see Supplemental Figure 2 online), demonstrating that they could recognize the corresponding epitopes after tissue processing. Conversely, labeling was not observed in either PSVs or ER protein bodies when the primary antibodies were omitted (see Supplemental Figures 2H and 2I online) or when control antibodies against Arabidopsis thaliana 2S albumins were used (see Supplemental Figures 2J and 2K online).
Figure 2.
Localization of Storage Proteins in Aleurone PSVs.
(A) Electron micrograph of an aleurone cell at 22 DAP containing lipid bodies (LB) and PSVs with large inclusions (asterisks). CW, cell wall.
(B) to (H) Immunogold labeling of aleurone cells at 22 DAP ([B], [D], and [F]) and 18 DAP ([C], [E], [G], and [H]) with antibodies against maize storage proteins. Most of the signal was detected on PSV inclusions (arrowheads). G, Golgi; IM, intravacuolar membranes.
Bars = 500 nm.
Structural Analysis of Starchy Endosperm and Aleurone Cells in Maize at 22 DAP
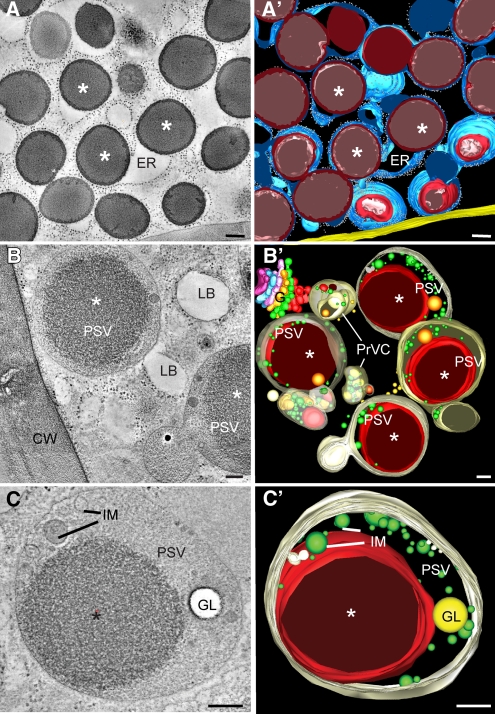
To understand how zeins and other storage proteins are transported to the PSVs in aleurone cells, we analyzed the architecture of the endomembrane system in the maize endosperm by dual-axis electron tomography. From six representative tomograms generated from high-pressure frozen/freeze-substituted maize endosperm samples at 22 DAP, we found protein bodies enclosed in a continuous ER network in the starchy endosperm (Figures 3A and 3A’; see Supplemental Movie 1 online). Although there were vacuoles in the developing starchy endosperm, we did not find evidence for storage protein–containing vacuolar compartments in these cells.
Figure 3.
Electron Tomographic Analysis of Aleurone and Starchy Endosperm Cells at 22 DAP.
(A) Tomographic slice (4.3 nm thick) of a starchy endosperm cell containing multiple protein bodies (asterisks) within the ER. The corresponding tomographic reconstruction can be seen in Supplemental Movie 1 online.
(A’) Tomographic model derived from the tomogram depicted in (A).
(B) Tomographic slice (4.3 nm thick) of an aleurone cell. PSVs with large inclusions (asterisks) and lipid bodies (LB) are the predominant storage compartment in these cells. CW, cell wall.
(B’) Tomographic model derived from the tomogram shown in (B). For simplicity, only PSVs, prevacuolar compartments (PrVC), and a Golgi stack (G) are depicted. This is a serial tomogram reconstructed from four serial 250-nm-thick serial sections.
(C) and (C’) Tomographic slice and tomographic model of a PSV. Note the presence of a large inclusion (asterisk), intravacuolar membranes (IM), and a globoid (GL). The corresponding tomographic reconstruction can be seen in Supplemental Movie 2 online.
Bars = 100 nm.
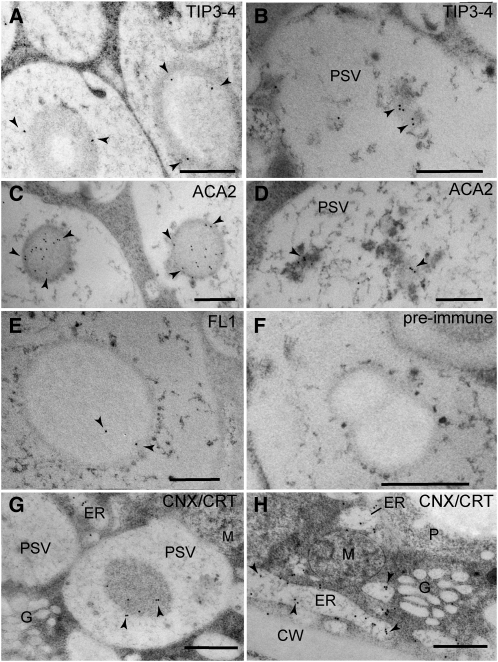
The architecture of the endomembrane system in aleurone cells was strikingly different, with PSVs and lipid bodies occupying most of the cellular volume (Figures 3B and 3B’). In addition to the zein-rich protein inclusions, the aleurone PSVs contained one or more globoids (crystals of phytic acid salts) and a large system of intravacuolar membranes (Figures 3C and 3C’; see Supplemental Movie 2 online). No discernable membrane was found surrounding the zein-rich inclusion within PSVs (Figure 3C; see Supplemental Movie 2 online). However, we detected proteins in the PSV inclusions and the intravacuolar membranes that are usually localized to the ER membrane, suggesting the presence of ER-type membranes within PSV inclusions. These ER markers included the aquaporin TIP3-4 (for Tonoplast Intrinsic Protein 3-4; Holding et al., 2007) (Figures 4A and 4B), ACA2 (for Arabidopsis Ca2+-ATPase, isoform 2; Harper et al., 1998) (Figures 4C and 4D; see Supplemental Figures 3A to 3C online), and FL1 (for Floury 1), which specifically decorates the ER membrane surrounding starchy endosperm protein bodies (Holding et al., 2007) (Figures 4E and 4F). In addition, the PSV matrix, protein inclusions, and intravacuolar membranes were labeled by antibodies that recognize the ER-resident proteins calnexin and calreticulin (Pagny et al., 2000), which further confirmed the accumulation of ER proteins in PSVs (Figure 4G). We also detected heavy labeling of calnexin/calreticulin on the aleurone ER as expected, but not on Golgi stacks (Figures 4G and 4H). In the starchy endosperm, the anticalnexin/calreticulin antibodies heavily labeled the ER protein bodies but not the vacuoles (see Supplemental Figure 3D online).
Figure 4.
Immunogold Detection of ER Proteins inside Aleurone PSVs at 18 DAP.
(A) and (B) TIP3-4 labeling on PSV inclusions (A) and PSV intravacuolar membranes (B).
(C) and (D) ACA2 detection on PSV inclusions (C) and PSV intravacuolar membranes (D).
(E) FL1 labeling on PSV inclusions.
(F) FL1 preimmune serum.
(G) and (H) calnexin/calreticulin labeling on PSV inclusions (G) and ER cisternae (H). Note the complete lack of labeling on Golgi stacks (G).
In all panels, arrowheads indicate the positions of gold particles. CW, cell wall; M, mitochondria P, plastid. Bars = 500 nm.
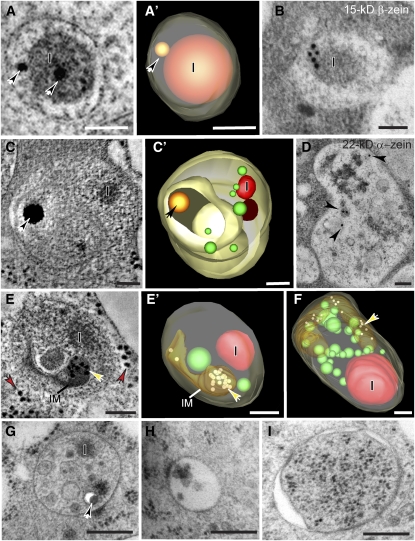
We also found several smaller membrane-bound compartments containing zeins (Figure 5). Some of them resembled small ER protein bodies. They were ~200 nm in diameter, were surrounded by a single membrane, and contained a central zein-rich aggregate often associated with one or more small globoids (Figures 5A, 5A’, and 5B). Other compartments were larger, ranging from 350 to 700 nm in diameter, with multiple layers of membranes and one or more zein-rich inclusions (Figures 5C to 5G). Most of these larger multilayered compartments also enclosed tubular membranes that contained structures that by size (18 nm) and electron density were interpreted to be ribosomes (Figures 5E, 5E’, and 5F, yellow arrows; see Supplemental Movies 3 and 4 online). Multivesicular bodies (MVBs) of ~250 nm in diameter (Figure 5H) and double-membrane autophagosome-like structures (Figure 5I) ranging from 0.8 to 1 μm in diameter could be seen in aleurone cells, but no zeins, α-globulin, or legumin-1 were evident in these compartments.
Figure 5.
Analysis of Zein-Containing Compartments, MVBs, and Putative Autophagosomes in Aleurone Cells.
(A) and (A') Tomographic slice (A) and derived tomographic model (A') of a membrane-bound compartment containing an aggregate or inclusion (I) identified as zeins (see [B]) and two electron-dense globoids (arrows).
(B) Immunodetection of 15-kD β-zein in an organelle similar to the one depicted in (A) and (A').
(C) to (F) Tomographic reconstructions of zein-containing prevacuolar compartments. Tomographic slice (C) and derived tomographic model (C') of an organelle consisting of several layers of concentric membranes, with aggregates or inclusions (I) identified as zeins (see [D]), and an electron-dense globoid (arrow).
(D) Immunodetection of 22-kD α-zein in an organelle similar to the ones depicted in (C), (C'), (E), (E'), (F), and (G).
(E) and (E’) Tomographic slice (E) and corresponding tomographic model (E’) of a prevacuolar compartment containing internal membranes (IM) and enclosed ribosomes (yellow arrows). For direct comparison, some ribosomes located in the cytoplasm are indicated by red arrows in (E). The corresponding tomographic reconstruction can be seen in Supplemental Movie 3 online.
(F) Tomographic model of another zein-containing prevacuolar compartment. The corresponding three-dimensional rendition of this model can be seen in Supplemental Movie 4 online.
(G) to (I) Electron micrographs of three different types of membrane-bound organelles that are likely involved in transport to PSVs in aleurone cells. Prevacuolar compartment containing an inclusion (I) and a globoid (arrow) (G); MVB (H); and autophagosome-like structure (I). For easy side-by-side comparison, all the images in (G) to (I) are depicted at the same magnification.
Bars = 100 nm in (A) to (F) and 200 nm in (G) to (I).
Younger Aleurone PSVs Contain Larger Intravacuolar Membrane Systems
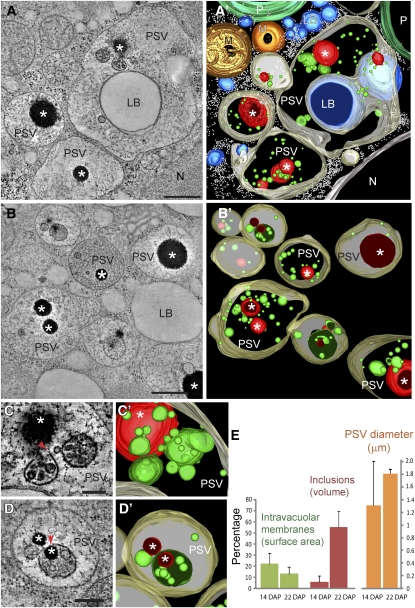
We also obtained electron tomograms of younger aleurone cells at 14 DAP. At this stage, the PSV intravacuolar membranes were more abundant and seemed to be preferentially associated with the zein-rich inclusions (Figures 6A to 6D). Some intravacuolar membranes formed multivesicular spherical structures, completely closed or partially open (Figures 6C, 6C’, 6D, and 6D’). In some cases, intravacuolar membranes were found to encircle, partially or completely, the zein-rich inclusions (Figures 6D and 6D’).
Figure 6.
Electron Tomographic Analysis of PSVs in Aleurone Cells at 14 DAP.
(A) to (B') Tomographic slices ([A] and [B]) and derived tomographic models ([A’] and [B’]) of aleurone cells showing the distribution of zein-rich inclusions (asterisks) and intravacuolar membranes (depicted in green) within the developing PSVs.
(C) to (D') Tomographic slices ([C] and [D]) and corresponding tomographic models ([C’] and [D’]) of intravacuolar membranes and zein-rich inclusions in PSVs. Note that some intravacuolar membranes form completely closed or partially open (arrows in [C] and [D]) multivesicular, spherical structures. In some cases, intravacuolar membranes enclose, partially or completely, the zein-rich inclusions ([D] and [D’]).
(E) Quantitative analysis of changes in intravacuolar surface area (expressed as a percentage of the total vacuolar membrane including the tonoplast), volume occupied by the zein-rich inclusions (expressed as a percentage of the total PSV volume), and PSV diameter between 14- and 22-DAP aleurone samples. Thirteen PSVs at 14 DAP and 12 PSVs at 22 DAP were considered in this analysis. The error bars denote the standard deviation.
LB, lipid body; M, mitochondrion; N, nucleus; P, plastid. Bars = 500 nm in (A) and (B) and 200 nm in (C) and (D).
To understand the dynamics of the intravacuolar membrane system and the zein-rich inclusions, we quantitated our tomographic reconstructions. For each PSV at either 14 or 22 DAP, we measured the total membrane surface area (surface area of tonoplast plus intravacuolar membranes) and calculated the percentage that corresponded to the intravacuolar membranes. We also calculated the percentage of the PSV total volume occupied by the zein-rich inclusions. Compared with PSVs at 14 DAP, the PSV size was much more uniform at 22 DAP (1.3 ± 0.7 μm at 14 DAP versus 1.8 ± 0.1 μm in diameter at 22 DAP; Figure 6E). The percentage of membrane surface area located in the vacuolar lumen changed from 22% (±10%) at 14 DAP to 13% (±6%) at 22 DAP. The zein-rich inclusions occupied on average 6% (±4%) of the PSV volume at 14 DAP and 55% (±14%) at 22 DAP.
Golgi-Processed Asn-Linked Glycans Can Be Detected in the PSV Matrix
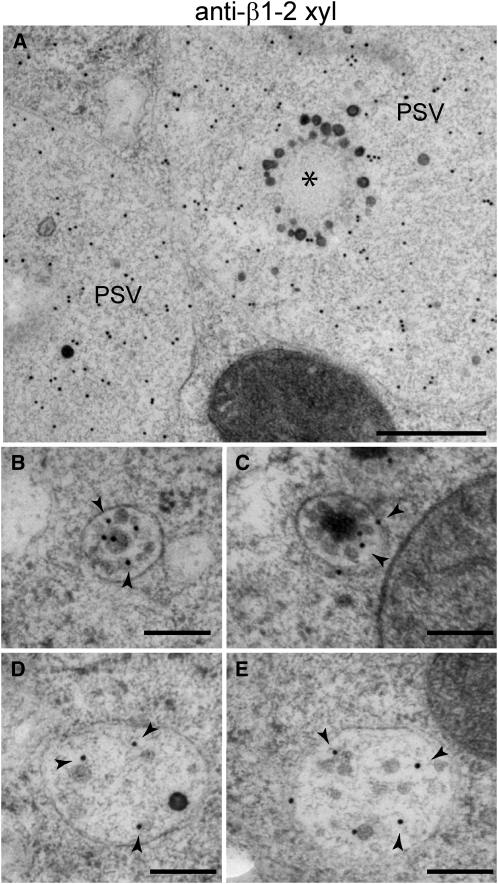
To determine the contribution of the Golgi to the transport of storage proteins to the PSV in aleurone cells, we probed 22-DAP endosperm samples for glycoproteins using antibodies against a β1-2 xylose epitope found on Golgi-processed Asn-linked glycans (Faye et al., 1993). Strong signals were detected in the PSV matrix and Golgi stacks but not in the PSV inclusions (Figure 7A; see Supplemental Figure 3E online). Consistently, no β1-2 xylose labeling was detected on ER protein bodies in the starchy endosperm (see Supplemental Figure 3F online), indicating that zeins, α-globulin, and legumin1 in both starchy endosperm and aleurone cells do not contain detectable Golgi-modified Asn-linked glycans.
Figure 7.
Immunodetection of β1-2 Xylose on Aleurone Cells at 22 DAP.
(A) β1-2 Xylose-positive labeling of PSV matrix. The PSV inclusion is indicated by an asterisk.
(B) and (C) β1-2 Xylose gold labeling (arrowheads) of MVBs.
(D) and (E) β1-2 Xylose gold labeling (arrowheads) of zein-containing prevacuolar compartments.
Bars = 500 nm in (A) and 200 nm in (B) to (E).
In the aleurone cells, we also detected strong β1-2 xylose signal on MVBs (86 gold particles/μm2 ± 43; n = 14 MVBs) and a weaker signal on the multilayered compartments that contain zeins (39 gold particles/μm2 ± 10; n = 12 compartments; labeling on cytoplasm was 1.0 gold particles/μm2 ± 0.8, n = 18 randomly selected 1-μm2 areas in 15 cells) (Figures 7B to 7E). This suggests that both MVBs and the larger zein-containing prevacuolar compartments are trafficking Golgi-processed glycoproteins.
FL1 Is Present in Aleurone Cells and Associates with Small ER Bodies
Our data suggest that zeins accumulate initially in the ER membranes of aleurone cells and then are delivered to multilayered prevacuolar compartments and PSVs. To understand why aleurone zeins do not form stable ER protein bodies like those in the starchy endosperm, we analyzed the behavior of FL1 (Figures 8A to 8G), a protein that localizes to ER protein body membranes and helps distribute zeins within ER protein bodies (Holding et al., 2007). We first confirmed the presence of FL1 in both aleurone and starchy endosperm tissues by immunoblot. Interestingly, FL1 abundance correlated with that of zeins (i.e., low in aleurone cells and much higher in the starchy endosperm) (Figure 8G).
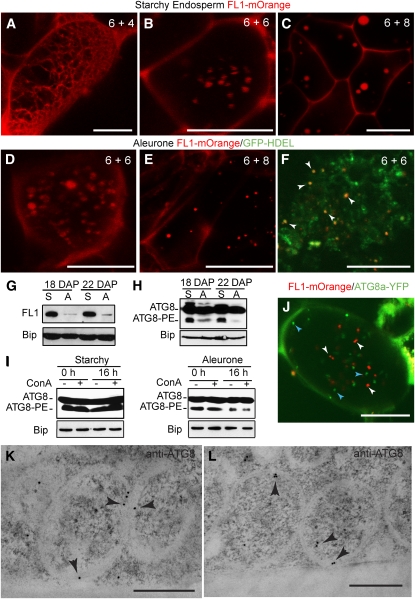
Figure 8.
FL1 and ATG8 Distribution in Aleurone and Starchy Endosperm Cells.
(A) to (C) Expression of FL1-mOrange in starchy endosperm cells of in vitro–cultured endosperms at different developmental stages: 6 DAP plus 4 DIC (A), 6 DAP plus 6 DIC (B), and 6 DAP plus 8 DIC (C). Note the change in the FL1-mOrange localization pattern, from a reticulated distribution in (A) to a punctuate pattern in (B) and (C).
(D) and (E) Expression of FL1-mOrange in aleurone cells of in vitro cultured endosperms at 6 DAP plus 6 DIC (D) and 6 DAP plus 8 DIC (E).
(F) Coexpression of FL1-mOrange and the ER marker GFP-KDEL in aleurone cells of in vitro–cultured endosperms at 6 DAP plus 8 DIC. Arrowheads indicate the localization of FL1-mOrange as discrete spherical structures in the ER.
(G) FL1 accumulation in starchy endosperm (S) and aleurone (A) samples at 18 and 22 DAP revealed by immunoblots. Bip was used as loading control.
(H) Profile of ATG8 proteins in starchy endosperm (S) and aleurone (A) samples at 18 and 22 DAP. Total protein extracts were subjected to SDS-PAGE in the presence of urea followed by immunodetection with antibodies against ATG8. Free (ATG8) and lipidated (ATG8-PE) forms were detected. Bip was used as loading control.
(I) Profile of ATG8 proteins in starchy endosperm and aleurone samples treated with ConA for 0 or 16 h. Bip was used as loading control.
(J) Coexpression of FL1-mOrange (white arrowheads) and ATG8a-YFP (blue arrowheads) in an aleurone cell of an in vitro–cultured endosperm at 6 DAP plus 8 DIC. Note the lack of colocalization of the two fluorescent signals.
(K) and (L) Immunogold detection of ATG8 (arrow heads) on double-membrane, autophagosome-like structures in starchy endosperm cells.
Bars = 500 nm in (K) and (L), 20 μm in (A) to (F), and 10 μm in (J).
To analyze the dynamics of FL1 during PSV and ER protein body assembly, we used a transgenic endosperm system, which can differentiate aleurone and starchy endosperm cells after isolation and culture (Gruis et al., 2006; Reyes et al., 2010). Endosperm tissue was excised from 6-DAP seeds and then transformed with Agrobacterium tumefaciens strains (Reyes et al., 2010) harboring a FL1pro:FL1-mOrange transgene either alone or together with the ER reporter OsAct1prom:GFP-HDEL (GFP for green fluorescent protein; Reyes et al., 2010). In this cultured system, aleurone and starchy endosperm cells show evidence of differentiation by 6 d in culture (DIC) and are completely differentiated by 8 to 10 DIC (Reyes et al., 2010). In both starchy endosperm and aleurone cells, FL1-mOrange changed in its localization from a reticulated ER distribution into a punctate pattern (Figures 8A to 8F). At 6 DIC, FL1-mOrange became more restricted to spherical bodies that likely represent ER protein bodies (Figures 8B to 8F), whereas at 8 DIC, FL1-mOrange-positive spherical bodies were clearly seen in both starchy endosperm (Figure 8C) and aleurone cells (Figure 8E).
ATG8 Is Not Associated with Zein-Containing Autophagic Membranes
The intravacuolar membranes and ER proteins inside the PSVs, the presence of multilayered prevacuolar compartments containing both zeins and cytoplasmic material, and the general lack of zein labeling in both Golgi stacks and MVBs are suggestive of a Golgi-independent, autophagic route for the vacuolar delivery of zeins. Since macroautophagy, the best-characterized autophagic mechanism for delivering cytoplasmic components to the vacuole, requires the lipidation of ATG8 (Klionsky et al., 2008; Rubinsztein et al., 2009; Chung et al., 2010), we assayed for this modification during aleurone development by immunodetection after urea-based SDS-PAGE separation. At both 18 and 22 DAP, we found consistently less lipidated ATG8 in aleurone-enriched fractions compared with the starchy endosperm–enriched fractions (Figure 8H). Because lipidated ATG8 is delivered to the vacuole and degraded during macroautophagy, the lower aleurone levels could reflect vacuolar breakdown. To test for this turnover, we treated aleurone peels and starchy endosperm with concanamycin A (ConA), a drug that interferes with vacuolar acidification and concomitantly stabilizes autophagic bodies in the vacuolar lumen (Yoshimoto et al., 2004; Thompson et al., 2005). ConA had no measurable effect on ATG8 lipidation (Figure 8I), suggesting that the lower levels of lipidated ATG8 in aleurone cells are not caused by vacuolar degradation.
To explore further the possible role of ATG8 in the delivery of zeins to the aleurone PSVs, we simultaneously transformed either FL1pro:FL1-mOrange or Ubpro:22-kD α−zein-mOrange together with the Ubpro:ATG8a-YFP (for yellow fluorescent protein) reporter into cultured endosperms (Figure 8J; see Supplemental Figure 4 online). ATG8a-YFP localized in both aleurone and starchy endosperm cells to ~1-μm cytoplasmic compartments could represent autophagophores or autophagosomes. These compartments did not appear to colocalize with either FL1-mOrange or 22-kD α-zein-mOrange in aleurone cells (Figure 8J; see Supplemental Figure 4 online).
Using anti-ATG8 immunogold labeling of cryofixed samples as an alternative, we also failed to detect ATG8 on either the cytoplasmic compartments containing zeins or PSV internal membranes. The anti-ATG8 antibodies did label the double membrane-bound cytoplasmic structures (Figures 8K and 8L) shown previously in Figure 5I, but these ATG8-positive structures did not appear to contain zein aggregates. The experiments together suggest that the vacuolar delivery of zeins in aleurone cells does not proceed via a typical ATG8-dependent macroautophagic process.
DISCUSSION
Although Zein Genes Are Transcribed in Aleurone Cells, Zein Proteins Accumulate in Large Quantities Only in the Starchy Endosperm
The expression and accumulation pattern of zeins and other storage proteins have been extensively studied in the maize starchy endosperm (Marks et al., 1985; Lending and Larkins, 1989; Or et al., 1993; Wang and Messing, 1998; Woo et al., 2001; Shewry and Halford, 2002; Marzábal et al., 2008; Arcalis et al., 2010). Despite the importance of the aleurone layer for endosperm development and seed germination, the expression and trafficking of storage proteins in this tissue has not been analyzed in detail. We show that although only the starchy endosperm accumulates significant amounts of zeins and other storage proteins, aleurone cells also produce abundant zein transcripts (Figure 1A). It is unclear why the aleurone cells actively transcribe zein genes if they are not important sites of zein accumulation. A similar situation has been reported for wheat prolamin. In that case, the α-gliadin promoter is active in both aleurone and starchy endosperm cells, but the encoded protein can be detected only in the starchy endosperm cells (Van Herpen et al., 2008).
One possible reason for this is that zeins do not form stable ER protein bodies in aleurone cells. Zein mRNAs could be actively translated and the resulting polypeptides immediately degraded, or the mRNAs could be translated at very low rates. Transcripts of prolamins in the rice (Oryza sativa) starchy endosperm have been shown to be recruited to specific ER domains where they are translated and the corresponding protein translocated to form to ER protein bodies (Choi et al., 2000; Hamada et al., 2003; Crofts et al., 2010). Although it is a matter of debate whether this is also the case for maize prolamin transcripts (Kim et al., 2002; Washida et al., 2004), proteins required for the efficient accumulation of zein aggregates inside the ER may not be expressed in aleurone cells. This scenario could explain the small sizes of the protein bodies observed in aleurone cells (Figure 8E). In fact, FL1, a protein important for proper localization of the 22-kD α-zeins within starchy endosperm ER protein bodies, is expressed at much lower levels in the aleurone cells (Figure 8G). Given that fl1 mutants are able to form protein bodies in the starchy endosperm, it is possible that other uncharacterized proteins important for prolamin protein body formation are not present in sufficient amounts in the aleurone cells.
The scarce accumulation of zeins in aleurone cells could also be related to differences in zein composition compared with the starchy endosperm. It is well known that when the ratio of zein polypeptides and/or the temporal accumulation of zeins are altered, the morphology and size of starchy endosperm ER protein bodies are subsequently disturbed, presumably by affecting protein aggregation/packing (Zhang and Boston, 1992; Coleman et al., 1997; Kim et al., 2006; Wu et al., 2010; Wu and Messing, 2010). The N-terminal domain of γ-zein has been shown to interact with liposomes (Kogan et al., 2004), implying that this domain could interact with ER membranes and contributes to prolamin retention in the ER. The fact that the 27-kD γ-zein, which is thought to be essential for protein body initiation (Lending and Larkins, 1989), is hardly detected in aleurone cells may also explain the limited accumulation of other zeins in aleurone cells. Alternatively, it is possible that only the starchy endosperm cells, which will undergo programmed cell death, can commit such a large proportion of their ER to the accumulation of storage proteins. Metabolically active cells, such as aleurone cells, may instead need to sequester protein stores in vacuoles outside the ER to retain viability.
Aleurone PSVs Contain Inclusions of Zeins, α-Globulin, and Legumin-1
The presence of large inclusions within aleurone PSVs in maize has been described previously (Kyle and Styles, 1977), but their composition was unknown. We show here that these inclusions are rich in zeins, α-globulin, and legumin-1, which in the starchy endosperm accumulate in ER protein bodies. When protein inclusions inside PSVs appear as a crystalline lattice, they are called crystalloids. Crystalloids have been described in rice and in seeds of some dicots, such as tomato (Solanum lycopersicum) and pumpkin (Cucurbita maxima; Krishnan et al., 1986; Krishnan and White, 1995; Kumamaru et al., 2010). Besides storage proteins, crystalloids contain integral membrane proteins and lipids, which have been hypothesized to form a multilamellar membrane/lattice structure (Jiang et al., 2000; Gillespie et al., 2005; Oufattole et al., 2005). However, our data suggest that the organization of the maize aleurone PSVs differs from the one reported for crystalloid-containing PSVs in dicots. First, the inclusions inside the aleurone PSVs do not have a crystalline lattice arrangement and thus should not be considered crystalloids. Second, the intravacuolar membrane system in maize aleurone PSVs (this study) and PSVs in wheat and barley (Bechtel et al., 1982b; Rechinger et al., 1993) is not exclusively associated with the zein-containing inclusion but extends throughout the PSV matrix.
Zeins Are Transported to the Aleurone PSVs via a Golgi-Independent Pathway
Another curious observation in our study is the fact that zeins do not remain as ER protein bodies in the cytoplasm of aleurone cells but are delivered to PSVs. Examples exist of prolamins trafficked to the PSVs in the starchy endosperm of other cereals such as wheat and barley (Levanony et al., 1992; Rechinger et al., 1993) and vacuolar delivery of zeins heterologously expressed in dicots (Coleman et al., 2004). Here, we found two completely different trafficking routes for the same storage proteins in two adjacent cell types within the same tissue.
In maize aleurone cells, we found evidence that zeins, α-globulin, and legumin-1 are trafficked directly from the ER to the PSV without transiting through the Golgi. First, we were unable to detect these storage proteins on Golgi stacks in multiple immunogold labeling experiments. Second, we could not detect Golgi-processed complex glycans in the PSV inclusion where zeins, α-globulin, and legumin-1 are localized. Third, ER-resident proteins such as calnexin/calreticulin, ACA2, FL1, and TIP3-4 could be found inside PSVs but not in Golgi stacks, suggesting a direct ER-to-PSV pathway in aleurone cells.
Hordeins (barley prolamins) and gliadin and glutenins (wheat prolamins) are synthesized in the ER and delivered to PSVs structurally similar to the ones described here in the maize aleurone. A Golgi-dependent route for trafficking of wheat and barley prolamins has been proposed based on the immunogold detection of prolamins in Golgi-associated vesicles (Bechtel et al., 1982a; Møgelsvang and Simpson, 1998). However, in wheat, a Golgi-independent pathway seems to transport the bulk of the prolamins to the vacuole (Levanony et al., 1992; Shy et al., 2001).
Autophagy in Cereal Endosperm Cells
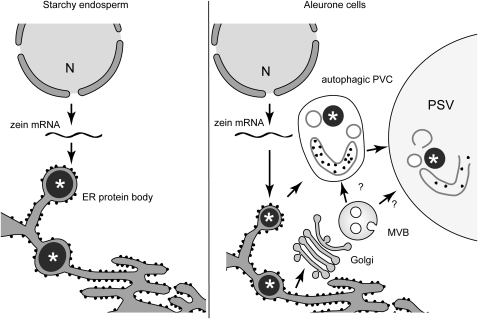
Based on electron microscopy (EM) imaging of chemically fixed samples, Levanony et al. (1992) suggested that the Golgi-independent transport of prolamins in wheat endosperm involves de novo formation of PSVs. Electron-lucent vesicles would coalesce around prolamin ER bodies, giving rise to vacuoles with prolamin inclusions. It is difficult to determine the origin of their electron-lucent structures due to of the lack of organelle markers and the difficulty in establishing connectivity between organelles in two-dimensional EM images. Our tomographic analysis indicates that zeins first accumulate in the ER and then are sequestered into complex prevacuolar compartments that contain internal membranes, Golgi-processed glycans, and cytoplasmic material (ribosomes). The presence of ribosomes enclosed in large tubular structures within the multilamellar prevacuolar compartments suggests that they originate at least partially by autophagy. The presence of Golgi-processed complex glycans indicates that they also traffic Golgi cargo. When these compartments fuse with the PSV, this compartment not only delivers storage proteins and globoids, but also the intravacuolar membrane system observed in PSVs (Figure 9).
Figure 9.
Model Depicting the Mechanisms of Accumulation and Transport of Zeins in Starchy Endosperm and Aleurone Cells.
In starchy endosperm cells (left), zeins are synthesized and stored in the ER as protein bodies (asterisk). In aleurone cells (right), zeins first accumulate in the ER as protein aggregates (asterisk) and then are sequestered into complex prevacuolar compartments (PVC) that fuse with the PSV. The presence of internal membranes and ribosomes inside the prevacuolar compartments suggest that these compartments are originating, at least partially, by autophagy. The glycoproteins found in the PSV matrix traffic through a typical ER-Golgi-MVB pathway, although it is possible that the MVBs carrying these glycoproteins fuse with the autophagic PVCs before reaching the PSV.
A wide range of prevacuolar compartments containing seed storage proteins have been described. In dicots such as legumes and Arabidopsis, MVBs receive seed storage proteins and their processing proteases from the Golgi and carry them to the PSVs (Hohl et al., 1996; Robinson et al., 1998; Robinson and Hinz, 1999; Hillmer et al., 2001; Jolliffe et al., 2005; Otegui et al., 2006; Hinz et al., 2007). In developing pumpkin seeds, the pro-11S globulin and pro-2S albumin precursors segregate from the ER to form the central core of precursor-accumulating (PAC) vesicles, bypassing the Golgi (Hara-Nishimura et al., 1985, 1998, 2004; Fukasawa et al., 1988; Herman and Schmidt, 2004). PAC vesicles are between 200 and 400 nm in diameter and consist of an electron-lucent outer region with internal vesicles and an inner core of storage protein aggregates. Rice contains compartments similar to PAC vesicles with a central core of storage proteins (glutelins and α-globulins in this case) and an outer translucent layer with vesicles, but they differ from PAC vesicles in being larger (~1 μm in diameter) and having ribosomes decorating their surface (Takahashi et al., 2005). In addition, physical connections between these compartments and the ER have been suggested based on EM images (Takahashi et al., 2005).
It is hard to assess how all these prevacuolar compartments relate to each other. The zein-containing prevacuolar compartments in maize aleurone cells described here appear unique in being carriers for prolamins and having multiple layers of membranes and engulfed cytoplasmic material. Although these bodies display the hallmarks of autophagic compartments, they are neither surrounded by a double membrane nor decorated by ATG8, suggesting that they are not typical autophagosomes.
Although autophagosomes resulting from ATG8-dependent macroautophagy are the best-characterized autophagic organelles, there are still many aspects of their dynamics that are unclear. One is the origin of the autophagosomal membrane itself. Recent contributions have demonstrated that the autophagosomal membrane can be derived from mitochondria, ER, or even the plasma membrane depending on the cell types and autophagic trigger (Hayashi-Nishino et al., 2009; Cuervo, 2010; Ravikumar et al., 2010). Based on the recent identification in mammalian cells of an ATG8-independent route (Nishida et al., 2009), it is also possible that plants have alternative autophagic routes that remain invisible to ATG8-based reporters. Consequently, it is possible that the one described here in maize aleurone cells represents a specialized case of selective autophagy used to deliver storage protein aggregates from the ER to the PSV. The fact that these prevacuolar compartments contained Golgi-modified complex glycans suggests that they receive cargo from the Golgi. As autophagosomes in mammalian cells, the prolamin-containing compartments in aleurone cells, could eventually fuse with MVBs to form a hybrid prevacuolar compartment (Berg et al., 1998; Fader et al., 2008).
This unusual autophagic mechanism for the vacuolar delivery of prolamins may be unique in terms of being a specialized macroautophagy event but could be common to other cereal endosperms. The fact that the prolamin-accumulating PSVs in wheat and barley starchy endosperm cells not only contain a large intravacuolar membrane system like the one present in maize aleurone PSVs but also the ER chaperone BIP in the case of wheat, would suggest a similar origin for all these PSVs. Thus, the trafficking pathway of zeins in the maize aleurone cells may be a widespread mechanism for vacuolar delivery of prolamins in cereals. Clearly the application of genetics to this autophagic-like process is needed to resolve this route. Unfortunately, both the absence of strong maize atg mutant alleles and the large number of ATG paralogs in maize (Chung et al., 2009) makes this analysis challenging at present.
METHODS
Plant Material and Endosperm in Vitro Culture
Maize (Zea mays, inbred lines B73 and A636) were grown in a greenhouse under a 14-h-light/10-h-dark photoperiod, supplemental lighting (700 μmol m−2 s−1), and average temperature of 28°C during the day and 21°C at night. Endosperms from A636 inbred plants were dissected and cultured in vitro as described by Gruis et al. (2006) and Reyes et al. (2010). Briefly, developing endosperms at 6 DAP were excised from kernels and immediately placed on liquid culture medium (4.3 g/L Murashige and Skoog media, 0.5% [v/v] Murashige and Skoog vitamin stock solution, 5 mg/L thiamine HCl, 400 mg/L Asn, 10 μg/L 6-benzylaminopurine, and 15% sucrose, pH 5.8) until cocultivation with Agrobacterium tumefaciens. After the cocultivation, the isolated endosperms were placed on solid culture medium (liquid culture medium plus 3 g/L Gelrite) supplemented with 500 μg/mL carbenicillin and kept in the dark at 25°C.
Plasmids
A DNA fragment containing Fl1pro:Fl1-mOrange was cloned into the pTF101.1 vector (Frame et al., 2002). The Fl1pro:Fl1-mOrange construct was generated as following: the cDNA of mOrange fluorescent protein was amplified using the primers 5′-GAGCTCATGGTGAGCAAGGGC-3′ and 5′-GAATTCTCAGACTTGTACAGCTCGTC-3′ and cloned in the vector pTF101.1 using the restriction sites SacI and EcoRI (underlined in the primer sequences). The Fl1 gene was amplified using the primers 5′-AAACTCTAGACAATAGGGCTTCCTTGCTTTGATGTAGG-3′ (underlined XbaI) for the upstream promoter region and 5′-AACCGAGCTCCTTCTTATCTCTGTCATCCTCGCTTGTC-3′ (underlined SacI) for the end of the open reading frame without the stop codon.
The maize Ubiquitin-1 promoter and maize Atg8a (Chung et al., 2009) were cloned in the pMcG1005-YFP-Gateway vector (kindly provided by Karen McGinnis at the Florida State University). The mOrange cDNA (Shaner et al., 2004) was kindly provided by Roger Tsien (University of California–San Diego). The OsActin1pro:GFP-KDEL construct has been described elsewhere (Reyes et al. 2010). All the plasmids were introduced into competent A. tumefaciens cells EHA101 by freeze-thaw transformation (Chen et al., 1994).
RNA Extraction and Quantitative PCR
Frozen kernels were defrosted and the aleurone layer was immediately excised by mechanical peeling. For a preliminary determination of the levels of contamination with starchy endosperm cells, aleurone peels were incubated for 1 to 2 min with aqueous solution of 0.5% Nile blue (for lipid detection) (Macron Chemicals) or 1% KI and 1% iodine (for starch detection) (Electron Microscopy Sciences). Afterwards, the samples were rinsed several times with distillated water and observed using an Olympus BX60 epifluorescence microscope (Olympus America). Total RNA was extracted as described by Holding et al. (2007). Briefly, both starchy endosperm tissue and aleurone peels were refrozen in liquid nitrogen and ground in NTES buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, and 1% SDS) to avoid starch solubilization. Nucleic acids were extracted three times using Tris-buffered phenol/chloroform (1:1), pH 8, followed by 10,000g centrifugation. The RNA present in the aqueous phase was later purified using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA was reversely transcribed to the first strand of the cDNA using oligo(dT) in a 20-μL reaction volume using AMV transcriptase (Promega). For RT-PCR, 1 μL of RT product was used as PCR template in a 20-μL volume reaction, and different PCR cycles were conducted to optimize results for each gene. PCR products were separated in 1.5% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light.
For quantitative RT-PCR experiments, cDNAs were amplified using Brilliant II SYBR Green Q-PCR Mastermix (Stratagene) in an Mx3000P real-time PCR instrument (Stratagene). Relative gene expression data were generated by the comparative Ct method (Pfaffl, 2004; Schmittgen and Livak, 2008) using the Ubc9 gene as reference. All of the quantification experiments were done with two biological replicates in triplicate. All the primers were validated, and only those with efficiencies between 90 and 100% were used for quantification experiments. All of the primers used for quantitative and/or RT-PCR are listed in the Supplemental Table 1 online.
EM and Immunogold Labeling
Thin slices of endosperm tissue from developing kernels at 14, 18, and 22 DAP were processed by high-pressure freezing/freeze substitution. Pieces of tissue were transferred to freezing planchettes containing 0.1 M sucrose and high-pressure frozen in a Baltec HPM 010 unit (Technotrade). Substitution was performed in 2% OsO4 in anhydrous acetone at –80°C for 72 h, followed by slow warming to room temperature over a period of 2 d. After several acetone rinses, samples were removed from the holders and infiltrated in Epon resin (Ted Pella) according to the following schedule: 5% resin in acetone (4 h), 10% resin (12 h), 25% resin (12 h), and 50, 75, and 100% resin (24 h for each concentration). Polymerization was performed at 60°C. Sections were stained with 2% uranyl acetate in 70% methanol for 10 min, followed by Reynold’s lead citrate (2.6% lead nitrate and 3.5% sodium citrate, pH 12.0), and observed in a FEI CM120 electron microscope.
For gold immunolabeling, high-pressure frozen samples were substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) and 0.2% glutaraldehyde (Electron Microscopy Sciences) in acetone at −80°C for 72 h and warmed to −50°C for 24 h. After several acetone rinses, these samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences) for 72 h and polymerized at −50°C under UV light for 48 h. Sections were mounted on formvar-coated nickel grids and blocked for 20 min with a 10% (w/v) solution of nonfat milk in Tris-buffered saline (TBS) containing 0.1% Tween 20. The sections were incubated with the primary antibodies (1:10 in TBS-Tween 20) for 1 h, rinsed in TBS containing 0.5% Tween 20, and then transferred to the secondary antibody (anti-rabbit IgG 1:10) conjugated to 15-nm gold particles for 1 h. Controls omitted the primary antibodies. All the antibodies have been characterized elsewhere: TIP3-4 and FL1 (Holding et al., 2007), ACA2 (Harper et al., 1998), calnexin-calreticulin (Pagny et al., 2000), α-globulin, legumin-1, 22-kD α-, 19-kD α-, 15-kD β-, 18-kD δ-, and 27-kD γ-zeins (Woo et al., 2001), 2S albumin (Scarafoni et al., 2001), and β1-2 xylose (Faye et al., 1993).
Electron Tomography
Semithick sections (250 nm) from Epon-embedded endosperm samples at 14 and 22 DAP were imaged in a FEI Tecnai TF30 300-kV intermediate-voltage electron microscope as described by Otegui et al. (2006). Images were taken between 60° and −60°, at 1° intervals about two orthogonal axes (Mastronarde, 1997), and collected in a Gatan digital camera at a pixel size of 1.0 nm. Tomograms were computed for each set of aligned tilts using the R-weighted back projection algorithm (Gilbert, 1972). Merging of the two single-axis tomograms into a dual-axis tomogram involved a warping procedure (Mastronarde, 1997). Dual axis electron tomograms were calculated, displayed, and segmented using the IMOD package (Kremer et al., 1996). The thinning factor for each tomogram was calculated and corrected for in the models. Quantitative analysis of membrane surface area and volumes was performed using the imodinfo program of the IMOD package (http://bio3d.colorado.edu/imod/).
Immunoblot Analysis
Proteins from starchy endosperm samples or aleurone peels were extracted as described by Leiva-Neto et al. (2004). Briefly, the samples were ground with a plastic pestle in NETN extraction buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20 mM Na2EDTA, 0.5% Triton X-100, 5 mM NaF, 1 mM Na orthovanadate, freshly added 1 mM phenylmethylsulfonyl fluoride, and 1 mM DTT) and protease inhibitor cocktail (Sigma-Aldrich). The debris was removed by centrifugation at 14,000g for 10 min. Protein extracts were diluted 1:10 in extraction buffer, and protein concentration was determinate using Bradford assay (Bio-Rad). Equal amount of proteins were separated by 12% (w/v) SDS polyacrylamide gel and then transferred to a cellulose membrane in a submerged blotting system (Bio-Rad). For the detection of the 2S albumin, total protein extracts from Arabidopsis thaliana seeds, starchy endosperm, and aleurone fractions were separated by Tricine-SDS-PAGE (16% acrylamide plus bisacrylamide, 3% cross-linker, and 6M urea) as described by Schägger (2006). Membranes were blocked for 1 h with TBS containing 10% (w/v) nonfat dry milk. All antibody solutions were prepared in TBS containing 0.05% (v/v) Tween 20. Proteins that cross-reacted with antibodies were detected with chemiluminescent substrates (Pierce) and visualized on film. The antibodies were diluted as follows: FL1 (Holding et al., 2007) 1:1000; ACA2 (Harper et al., 1998) 1:2000; Bip (Hsc70, monoclonal antibody [1D9]; Stressgen) 1:1000; 15-kD β-zein 1:2000; 18-kD δ-zein 1:3500; 27-kD γ-zein 1:5000; α-globulin 1:3000; legumin-1 1:3000 (Woo et al., 2001), and 19- and 22-kD α-zein 1:1000 (kindly provided by B.A. Larkins). The 2S albumin large chain antibody (Scarafoni et al., 2001) was used in a 1:300 dilution, and it was kindly donated by A. Scarafoni.
ATG8 Lipidation Analysis
The lipidation analysis of ATG8 and the ConA treatment were done as described by Chung et al. (2009, 2010). Briefly, total proteins were extracted using SDS-PAGE sample buffer from equal fresh weight of starchy endosperm and aleurone enriched fractions. The samples were then subjected to SDS-PAGE on 12% polyacrylamyde gel with 6 M urea. Both free ATG8 and the ATG8-PE adduct were detected by immunoblot analysis with anti-ATG8 antibodies (Thompson et al., 2005). For ConA treatment, fresh starchy and aleurone fractions were incubated with 1 μM ConA (Wako Chemicals) or an equivalent volume of DMSO for 16 h before total protein extraction.
Confocal Imaging of Fluorescent Proteins
Cross or paradermal sections of the transformed endosperms were imaged using a 510 Zeiss laser scanning confocal microscope. GFP and YFP were excited with a 488-nm excitation line; GFP emission was detected using a 500- to 530-nm band-pass filter, whereas YFP emission was detected with a 500- to 550-nm IR band-pass filter. mOrange was excited with a 514-nm excitation line, and the emission was collected using a 550- to 590-nm band-pass filter. The multitrack mode was used for sequential imaging of GFP and mOrange or YFP and mOrange. The emission spectra of GFP, YFP, and mOrange were confirmed for every image using the spectral Meta detector. The images were analyzed using the LSM image browser (www.zeiss.com/lsm) and edited using Adobe Photoshop CS4.
Accession Numbers
Sequence data for the genes used in this article can be found in the GenBank/EMBL data libraries under accession numbers AF371266 (10-kD δ-zein), AF371264 (15-kD β-zein), AF371262 (16-kD γ-zein), AF371265 (18-kD δ-zein), AF371268 (19-kD α-zein D2), AF371274 (22-kD α-zein Z1), AF371261 (27-kD γ-zein), AF371279 (50-kD γ-zein), AF371279 (legumin-1), AF371278 (α-globulin), AF034946.1 (Ubc9), NM_001112070.1 (Vp1), NM_001112419 (Bt1), EF536720 (Fl1), FJ445006 (Atg8a), and DQ141598 (Ubiquitin-1 promoter).
Supplemental Material
The following materials are available in the online version of this article.
Supplemental Figure 1. Aleurone Peels Excised from B73 Kernels and Stained with Nile Blue for Detection of Lipids and with IKI for Starch Detection.
Supplemental Figure 2. Immunogold Detection of Storage Proteins in Starchy Endosperm Cells and Negative Control Labeling on Aleurone PSVs.
Supplemental Figure 3. Additional Immunogold-Labeling Images with Anti-ACA2, Anti-Calnexin/Calreticulin, and Anti-β1-2 Xylose Antibodies and Immunoblot of ACA2 in Maize Protein Extracts.
Supplemental Figure 4. Coexpression of 22-kD α-Zein-mOrange and ATG8a-YFP in Aleurone Cells of an in Vitro–Cultured Endosperm at 6 DAP Plus 8 DIC.
Supplemental Table 1. Primers Used for Quantitative and/or RT-PCR.
Supplemental Movie 1. Tomographic Slices of an Electron Tomogram from a Starchy Endosperm Cell Depicted in Figures 3A and 3A’.
Supplemental Movie 2. Tomographic Slices of an Electron Tomogram from an Aleurone PSV Depicted in Figures 3C and 3C’.
Supplemental Movie 3. Tomographic Slices of an Electron Tomogram from the Zein-Containing Prevacuolar Compartment Depicted in Figures 5E and 5E’.
Supplemental Movie 4. Tomographic Model of the Zein-Containing Prevacuolar Compartment Depicted in Figure 5F.
Acknowledgments
We thank Kan Wang (Iowa State University) for providing the A. tumefaciens EHA101 strain and the pTF101.1 binary vector, Ajay Gang (Cornell University) for providing the OsAct1 promoter, Rebecca Boston (North Carolina State University) for the antibodies against BIP and calnexin/calreticulin, Brian Larkins (Arizona University) for the antibodies against α-zeins, Karen McGinnis (Florida State Univeristy) for the pMcG1005-YFP-Gateway vector, and Roger Tsien (University of California–San Diego) for the gift of the mOrange cDNA. We also thank The Boulder Laboratory for three-dimensional electron microscopy of cells (University of Colorado–Boulder) and for the use of the FEI Tecnai TF30 300kV intermediate-voltage electron microscope and the Plant Imaging Center at the Department of Botany (University of Wisconsin–Madison) for the use of the Zeiss 510 Meta laser scanning microscope. We thank Thomas Haas and Chadwin Barnes for their assistance in manual tomogram segmentation. This project is supported by the National Research Initiative Competitive Grants 2008-35304-18672 and 2008-02545 from the USDA National Institute of Food and Agriculture to M.S.O. and to R.V., respectively.
References
- Alvarez I., Geli M.I., Pimentel E., Ludevid D., Torrent M. (1998). Lysine-rich gamma-zeins are secreted in transgenic Arabidopsis plants. Planta 205: 420–427 [DOI] [PubMed] [Google Scholar]
- Arcalis E., Stadlmann J., Marcel S., Drakakaki G., Winter V., Rodriguez J., Fischer R., Altmann F., Stoger E. (2010). The changing fate of a secretory glycoprotein in developing maize endosperm. Plant Physiol. 153: 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L.J., Yoshimoto K. (2006). Autophagy in development and stress responses of plants. Autophagy 2: 2–11 [DOI] [PubMed] [Google Scholar]
- Bechtel D.B., Gaines R.L., Pomeranz Y. (1982a). Early stages in wheat endosperm formation and protein body initiation. Ann. Bot. (Lond.) 50: 507–518 [Google Scholar]
- Bechtel D.B., Gaines R.L., Pomeranz Y. (1982b). Protein secretion in wheat endosperm. Formation of the matrix protein. Cereal Chem. 59: 336–343 [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. (2010). Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T.O., Fengsrud M., Strømhaug P.E., Berg T., Seglen P.O. (1998). Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273: 21883–21892 [DOI] [PubMed] [Google Scholar]
- Bethke P.C., Swanson S.J., Hillmer S., Jones R.L. (1998). From storage compartment to lytic organelle: The metamorphosis of the aleurone protein storage vacuole. Ann. Bot. (Lond.) 82: 399–412 [Google Scholar]
- Cao X., Costa L.M., Biderre-Petit C., Kbhaya B., Dey N., Perez P., McCarty D.R., Gutierrez-Marcos J.F., Becraft P.W. (2007). Abscisic acid and stress signals induce Viviparous1 expression in seed and vegetative tissues of maize. Plant Physiol. 143: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Nelson R.S., Sherwood J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–668, 670 [PubMed] [Google Scholar]
- Choi S.-B., Wang C., Muench D.G., Ozawa K., Franceschi V.R., Wu Y., Okita T.W. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Clore A.M., Ranch J.P., Higgins R., Lopes M.A., Larkins B.A. (1997). Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Herman E.M., Takasaki K., Larkins B.A. (1996). The maize gamma-zein sequesters alpha-zein and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 8: 2335–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Larkins B.A. (1999). The prolamins of maize. In Seed Proteins, Shewry P.R., Casey R., eds (Dordrecht, The Netherlands: Kluwer Academic Press; ), pp. 109–139 [Google Scholar]
- Coleman C.E., Yoho P.R., Escobar S., Ogawa M. (2004). The accumulation of α-zein in transgenic tobacco endosperm is stabilized by co-expression of β-zein. Plant Cell Physiol. 45: 864–871 [DOI] [PubMed] [Google Scholar]
- Crofts A.J., Crofts N., Whitelegge J.P., Okita T.W. (2010). Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231: 1261–1276 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M. (2010). The plasma membrane brings autophagosomes to life. Nat. Cell Biol. 12: 735–737 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I. (2008). Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9: 230–250 [DOI] [PubMed] [Google Scholar]
- Faye L., Gomord V., Fitchette-Lainé A.C., Chrispeels M.J. (1993). Affinity purification of antibodies specific for Asn-linked glycans containing α 1—>3 fucose or β 1—>2 xylose. Anal. Biochem. 209: 104–108 [DOI] [PubMed] [Google Scholar]
- Filner P., Varner J.E. (1967). A test for de novo synthesis of enzymes: density labeling with H2O18 of barley alpha-amylase induced by gibberellic acid. Proc. Natl. Acad. Sci. USA 58: 1520–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame B.R., Shou H., Chikwamba R.K., Zhang Z., Xiang C., Fonger T.M., Pegg S.E.K., Li B., Nettleton D.S., Pei D., Wang K. (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa T., Hara-Nishimura I., Nishimura M. (1988). Biosynthesis, intracellular transport and in vitro processing of 11S globulin precursor proteins of developing castor bean endosperm. Plant Cell Physiol. 29: 339–345 [Google Scholar]
- Galili G., Altschuler Y., Levanony H. (1993). Assembly and transport of seed storage proteins. Trends Cell Biol. 3: 437–442 [DOI] [PubMed] [Google Scholar]
- Geng J., Klionsky D.J. (2008). The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.F.C. (1972). The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc. R. Soc. Lond. B Biol. Sci. 182: 89–102 [DOI] [PubMed] [Google Scholar]
- Gillespie J., Rogers S.W., Deery M., Dupree P., Rogers J.C. (2005). A unique family of proteins associated with internalized membranes in protein storage vacuoles of the Brassicaceae. Plant J. 41: 429–441 [DOI] [PubMed] [Google Scholar]
- Gruis F., Guo H., Selinger D.A., Tian Q., Olsen O.-A. (2006). Surface position, and not signalling from surrounding maternal tissues, specifies aleurone epidermal cell fate in maize endosperm organ cultures. Plant Physiol. 141: 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Choi S.-B., Wang C., Singh S., Kawai N., Franceschi V.R., Okita T.W. (2003). The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Matsushima R., Shimada T., Nishimura M. (2004). Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiol. 136: 3435–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Nishimura M., Akazawa T. (1985). Biosynthesis and intracellular transport of 11S globulin in developing pumpkin cotyledons. Plant Physiol. 77: 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., I, Shimada T., Hatano K., Takeuchi Y., Nishimura M. (1998). Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.F., Hong B., Hwang I., Guo H.Q., Stoddard R., Huang J.F., Palmgren M.G., Sze H. (1998). A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J. Biol. Chem. 273: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Herman E., Schmidt M. (2004). Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies provides an alternate pathway for protein transfer to the vacuole. Plant Physiol. 136: 3440–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S., Movafeghi A., Robinson D.G., Hinz G. (2001). Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J. Cell Biol. 152: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G., Colanesi S., Hillmer S., Rogers J.C., Robinson D.G. (2007). Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos. Traffic 8: 1452–1464 [DOI] [PubMed] [Google Scholar]
- Hohl I., Robinson D.G., Chrispeels M.J., Hinz G. (1996). Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J. Cell Sci. 109: 2539–2550 [DOI] [PubMed] [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen J.V., Bush D.S., Sticher L., Jones R.L. (1988). Evidence for precursor forms of the low isoelectric point α-amylase isozymes secreted by barley aleurone cells. Plant Physiol. 88: 1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Phillips T.E., Rogers S.W., Rogers J.C. (2000). Biogenesis of the protein storage vacuole crystalloid. J. Cell Biol. 150: 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe N.A., Craddock C.P., Frigerio L. (2005). Pathways for protein transport to seed storage vacuoles. Biochem. Soc. Trans. 33: 1016–1018 [DOI] [PubMed] [Google Scholar]
- Kim C.S., Gibbon B.C., Gillikin J.W., Larkins B.A., Boston R.S., Jung R. (2006). The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response. Plant J. 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Kim C.S., Woo Ym Y.M., Clore A.M., Burnett R.J., Carneiro N.P., Larkins B.A. (2002). Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell 14: 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S., Leroch M., Huynen M.A., Wahl M., Neuhaus H.E., Tjaden J. (2007). Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 282: 22481–22491 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., et al. (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan M.J., López O., Cocera M., López-Iglesias C., De La Maza A., Giralt E. (2004). Exploring the interaction of the surfactant N-terminal domain of gamma-Zein with soybean phosphatidylcholine liposomes. Biopolymers 73: 258–268 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Krishnan H.B., Franceschi V.R., Okita T.W. (1986). Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta 169: 471–480 [DOI] [PubMed] [Google Scholar]
- Krishnan H.B., White J.A. (1995). Morphometric analysis of rice seed protein bodies (Implication for a significant contribution of prolamine to the total protein content of rice endosperm). Plant Physiol. 109: 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamaru T., Ogawa M., Satoh H., Okita T.W. (2007). Protein body biogenesis in cereal endosperms. Endosperm O.A., Olsen, (Berlin: Springer; ), pp. 141–158 [Google Scholar]
- Kumamaru T., Uemura Y., Inoue Y., Takemoto Y., Siddiqui S.U., Ogawa M., Hara-Nishimura I., Satoh H. (2010). Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 51: 38–46 [DOI] [PubMed] [Google Scholar]
- Kyle D.J., Styles E.D. (1977). Development of aleurone and sub-aleurone layears in maize. Planta 137: 185–193 [DOI] [PubMed] [Google Scholar]
- Leiva-Neto J.T., Grafi G., Sabelli P.A., Dante R.A., Woo Y.M., Maddock S., Gordon-Kamm W.J., Larkins B.A. (2004). A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16: 1854–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending C.R., Larkins B.A. (1989). Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanony H., Rubin R., Altschuler Y., Galili G. (1992). Evidence for a novel route of wheat storage proteins to vacuoles. J. Cell Biol. 119: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu Y., Zhang D.-Z., Gillikin J.W., Boston R.S., Franceschi V.R., Okita T.W. (1993). Rice prolamine protein body biogenesis: A BiP-mediated process. Science 262: 1054–1056 [DOI] [PubMed] [Google Scholar]
- Marks M.D., Lindell J.S., Larkins B.A. (1985). Quantitative analysis of the accumulation of Zein mRNA during maize endosperm development. J. Biol. Chem. 260: 16445–16450 [PubMed] [Google Scholar]
- Marzábal P., Gas E., Fontanet P., Vicente-Carbajosa J., Torrent M., Ludevid M.D. (2008). The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol. Biol. 67: 441–454 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. (1997). Dual-axis tomography: An approach with alignment methods that preserve resolution. J. Struct. Biol. 120: 343–352 [DOI] [PubMed] [Google Scholar]
- Møgelsvang S., Simpson D.J. (1998). Changes in the levels of seven proteins involved in polypeptide folding and transport during endosperm development of two barley genotypes differing in storage protein localisation. Plant Mol. Biol. 36: 541–552 [DOI] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658 [DOI] [PubMed] [Google Scholar]
- Olsen O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16(Suppl): S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E., Boyer S.K., Larkins B.A. (1993). opaque2 modifiers act post-transcriptionally and in a polar manner on gamma-zein gene expression in maize endosperm. Plant Cell 5: 1599–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Herder R., Schulze J.M., Jung R., Staehelin L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oufattole M., Park J.H., Poxleitner M., Jiang L., Rogers J.C. (2005). Selective membrane protein internalization accompanies movement from the endoplasmic reticulum to the protein storage vacuole pathway in Arabidopsis. Plant Cell 17: 3066–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagny S., Cabanes-Macheteau M., Gillikin J.W., Leborgne-Castel N., Lerouge P., Boston R.S., Faye L., Gomord V. (2000). Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12: 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2004). Quantification strategies in real-time PCR. In A-Z of Quantitative PCR, Bustin S.A., (La Jolla, CA: International University Line; ), pp. 87–112 [Google Scholar]
- Pompa A., Vitale A. (2006). Retention of a bean phaseolin/maize γ-Zein fusion in the endoplasmic reticulum depends on disulfide bond formation. Plant Cell 18: 2608–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall J.J., Sutton D.W., Hanson S.F., Kemp J.D. (2005). BiP and zein binding domains within the delta zein protein. Planta 221: 656–666 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. (2010). Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechinger K.B., Simpson D.J., Svendsen I., Cameron-Mills V. (1993). A role for γ 3 hordein in the transport and targeting of prolamin polypeptides to the vacuole of developing barley endosperm. Plant J. 4: 841–853 [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Sun B., Guo H., Gruis D.F., Otegui M.S. (2010). Agrobacterium tumefaciens-mediated transformation of maize endosperm as a tool to study endosperm cell biology. Plant Physiol. 153: 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Baeumer M., Hinz G., Hohl I. (1998). Vesicle transfer of storage proteins to the vacuole: The role of the Golgi apparatus and multivesicular bodies. J. Plant Physiol. 152: 659–666 [Google Scholar]
- Robinson D.G., Hinz G. (1999). Golgi-mediated transport of seed storage proteins. Seed Sci. Res. 9: 267–283 [Google Scholar]
- Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. (2009). In search of an “autophagomometer”. Autophagy 5: 585–589 [DOI] [PubMed] [Google Scholar]
- Scarafoni A., Carzaniga R., Harris N., Croy R.R. (2001). Manipulation of the napin primary structure alters its packaging and deposition in transgenic tobacco (Nicotiana tabacum L.) seeds. Plant Mol. Biol. 46: 727–739 [DOI] [PubMed] [Google Scholar]
- Schägger H. (2006). Tricine-SDS-PAGE. Nat. Protoc. 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shannon J.C., Pien F.-M., Cao H., Liu K.-C. (1998). Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP—glucose into amyloplasts of maize endosperms. Plant Physiol. 117: 1235–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P.R., Halford N.G. (2002). Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 53: 947–958 [DOI] [PubMed] [Google Scholar]
- Shy G., Ehler L., Herman E., Galili G. (2001). Expression patterns of genes encoding endomembrane proteins support a reduced function of the Golgi in wheat endosperm during the onset of storage protein deposition. J. Exp. Bot. 52: 2387–2388 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Saito Y., Kitagawa T., Morita S., Masumura T., Tanaka K. (2005). A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant Cell Physiol. 46: 245–249 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Vierstra R.D. (2005). Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Van Herpen T.W.J.M., Riley M., Sparks C., Jones H.D., Gritsch C., Dekking E.H., Hamer R.J., Bosch D., Salentijn E.M.J., Smulders M.J.M., Shewry P.R., Gilissen L.J.W.J. (2008). Detailed analysis of the expression of an alpha-gliadin promoter and the deposition of alpha-gliadin protein during wheat grain development. Ann. Bot. (Lond.) 102: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Ceriotti A. (2004). Protein quality control mechanisms and protein storage in the endoplasmic reticulum. A conflict of interests? Plant Physiol. 136: 3420–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Messing J. (1998). Modulation of gene expression by DNA-protein and protein-protein interactions in the promoter region of the zein multigene family. Gene 223: 333–345 [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Messing J., Esen A., Okita T.W. (2004). Asymmetric localization of seed storage protein RNAs to distinct subdomains of the endoplasmic reticulum in developing maize endosperm cells. Plant Cell Physiol. 45: 1830–1837 [DOI] [PubMed] [Google Scholar]
- Woo Y.-M., Hu D.W.-N., Larkins B.A., Jung R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Holding D.R., Messing J. (2010). λ-Zeins are essential for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. USA 107: 12810–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2010). RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 153: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D.J. (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T., Kato H., Kuroda S., Abe S., Davies E. (2003). Uncleaved legumin in developing maize endosperm: Identification, accumulation and putative subcellular localization. J. Exp. Bot. 54: 913–922 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Boston R.S. (1992). Increases in binding protein (BiP) accompany changes in protein body morphology in three high-lysine mutants of maize. Protoplasma 171: 142–152 [Google Scholar]