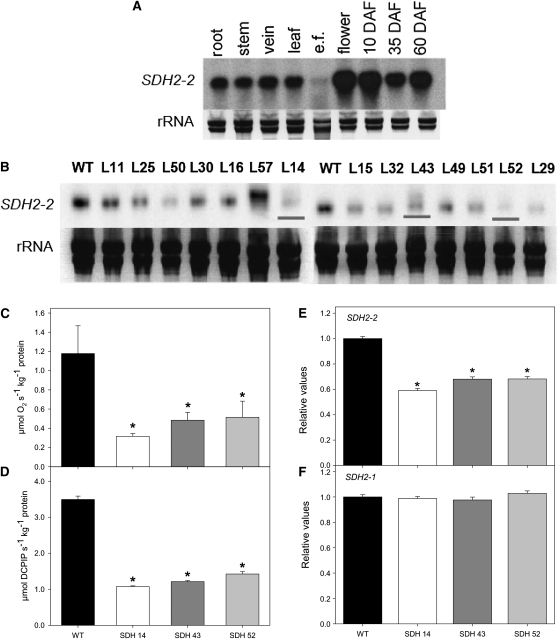
Figure 1.
Characterization and Expression of Tomato Succinate Dehydrogenase (SDH2-2).
(A) RNA gel blot containing total RNA extracted from different organs of tomato plants. Total RNA was obtained from root, stem, vein, leaf, epidermal fragments (e.f.), flowers, and fruits 10, 35, and 60 d after flowering (DAF).
(B) RNA gel blot analysis of leaves of 4-week-old transgenic tomato plants with altered expression of SDH2-2 compared with the wild type (WT). The full-length 825-bp cDNA encoding the iron-sulfur subunit of succinate dehydrogenase was cloned in the antisense orientation into the transformation vector pK2WG7 between the CaMV promoter and the ocs terminator (see Supplemental Figure 1B online), and 15 transgenic tomato plants were obtained. Screening of the lines (L) by RNA gel blot yielded three lines that displayed a considerable reduction of Sl SDH2-2 (shown in red lines).
(C) and (D) Succinate-dependent oxygen consumption in freshly isolated mitochondria of green fruits 35 DAF (C) and succinate-dependent DCPIP reduction determined in enriched mitochondria from tomato leaves (D). Data are presented as mean values ± se and are averages from three to five different mitochondrial isolations.
(E) and (F) Relative transcript abundance of mitochondrial Complex II subunits (SDH2-2 and SDH2-1, respectively). The abundance of SDH mRNAs was measured by qRT-PCR, and values are presented as mean ± se of six individual plants per line. Asterisks indicate values that were determined by the Student’s t test to be significantly different from the wild type (P < 0.05).