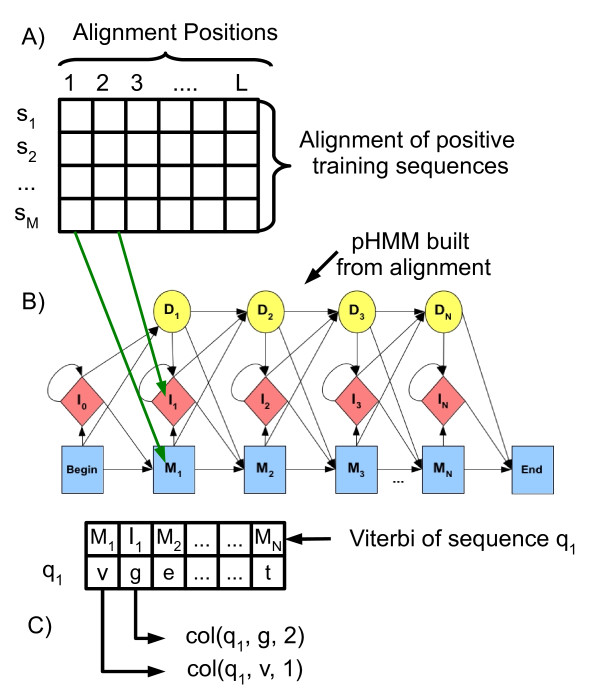
Figure 5.
Using pHMMs to create ground predicates for alignment positions. A) First, the positive training sequences are aligned. B) Second, a pHMM is built from this alignment. Each query sequence (negative training sequences, and positive and negative test sequences) is matched by pHMM producing the Viterbi output that shows the correspondence between each amino acid in the query sequence and pHMM states (match (M), insert (I) or delete (D)). C) Finally, we know the mapping between alignment positions and pHMM states, thus we create, for each query sequence, ground predicates in a similar way to Figure 4.