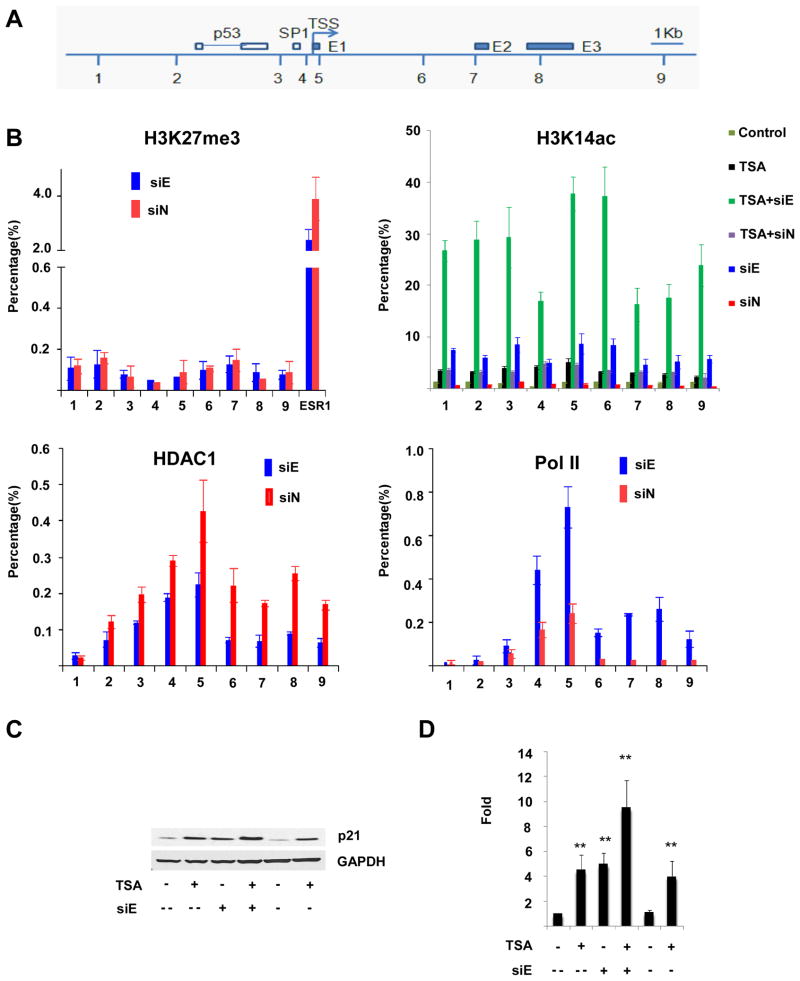
Figure 6.
Effect of EZH2 depletion on histone 3 interactions and modifications at CDKN1A. (A) Schematic drawing of CDKN1A locus depicting transcription start site (TSS), p53 and SP1 transcription factor binding sites, and the location of the three exons (E1,E2,E3). Positions of primer pairs used for chromatin immunoprecipitation (ChIP) are indicated (1–9). (B). ChIP analyses of CDKN1A using antibodies recognizing H3K27me3, H3K14ac, HDAC1, and RNA polymerase II were performed in UCD-Mel-N cells with EZH2 depletion (siE) or transfection with a control siRNA (siN). The precipitated DNA was amplified by real-time quantitative PCR (qPCR) using primers 1–9 from distinct regions of CDKN1A. Each experiment was repeated independently at least twice. The results were normalized to IgG control. The data are expressed as percentage of immunoprecipitated DNA relative to input DNA. Primers to the ESR1 gene were used as a positive control. Histone acetylation at CDKN1A was determined not only under the above conditions (siE and siN), but also in the above conditions with cells treated with 1 μM TSA (TSA+siE, TSA+siN), in cells treated with TSA alone (TSA), and in untreated or untransfected cells (Control). Error bars represent standard deviation from the mean. (C,D) Combined effects of HDAC inhibition and EZH2 depletion upon p21/CDKN1A transcript and protein expression. An additive effect of HDAC inhibition with trichostatin A (TSA) and EZH2 depletion (siE) on the upregulation of p21 protein expression as measured by Western blotting (C) and of CDKN1A mRNA expression as measured by quantitative real-time PCR (D) is observed relative to cells treated with TSA (TSA +) or EZH2 siRNA (siE +), respectively, alone. siE −, control siRNA cells; siE --, untreated cells; ** signifies p < 0.01 for indicated comparisons.