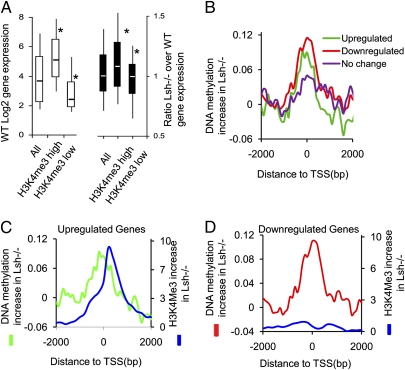
Fig. 4.
Relation of cytosine methylation changes and gene expression. (A) Box plot to demonstrate changes of gene expression changes for all genes (All) and those that were subgrouped based on changes in H3K4me3 and cytosine methylation. Genes were ranked according to their H3K4me3 changes in Lsh−/− MEFs (Fig 3D and Fig. S5B) from highest (H3K4me3 high) to lowest gain (H3K4me3 low, including those genes with H3K4me3 losses). (Left) Gene-expression level in WT MEFs according to the classification. (Right) Ratio of gene expression (Lsh−/− expression over WT expression), with values above 1 indicating up-regulation and below 1 indicating down-regulation. *P < 0.005. (B) Distribution of cytosine methylation changes in Lsh−/− MEFs at the 5′ end of genes that are twofold down-regulated in Lsh−/− MEFs (red, n = 581), twofold up-regulated in Lsh−/− MEFs (green, n = 320), or a control group without any change (purple, n = 543). (C) Distribution of cytosine methylation changes (green) and H3K4me3 changes (blue) in Lsh−/− MEFs at the 5′ end of genes that are twofold up-regulated in Lsh−/− MEFs. (D) Distribution of cytosine methylation changes (red) and H3K4me3 (blue) changes in Lsh−/− MEFs at the 5′ end of genes that are twofold down-regulated in Lsh−/− MEFs.