Figure 5.
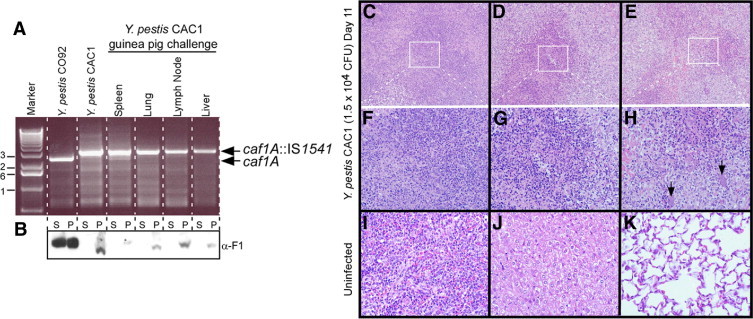
The Y. pestis caf1A::IS1541 variant, strain CAC1, can cause bubonic plague in guinea pigs without reverting to an F1+ phenotype. A: Genomic DNA isolated from Y. pestis CO92 (wild type), Y. pestis CAC1, or plague bacteria that were isolated from spleen, lung, lymph node, or liver tissues of a guinea pig that died of s.c. challenge with the CAC1 strain (asterisk in Figure 4B) was examined by PCR with oligonucleotide primers specific for the caf1A gene of pFRA. Arrows denote the mobility of PCR amplification products specific for caf1A or caf1A::IS1541. The 1-kb DNA ladder (Promega Corp, Madison, WI) was used as a calibration standard (marker). B:Y. pestis broth cultures from the same strains as described in A were centrifuged to separate proteins secreted into the culture supernatant (S) from the bacterial sediment (P) and analyzed by immunoblotting using polyclonal rabbit sera with antibodies specific for rF1 (α-F1). C–K: Spleen, liver, and lung tissues of guinea pigs were removed during necropsy, fixed in formalin, thin sectioned, and stained with H&E. The tissues of an uninfected guinea pig (I, J, and K) were compared with those of the animal that died of s.c. Y. pestis CAC1 challenge (asterisk in Figure 4B). The white boxes in panels C, D, and E identify the areas of enlargement in panels F, G, and H. Arrows denote bacterial microcolonies. Original magnification: ×200 (C–E); ×400 (F–K).
